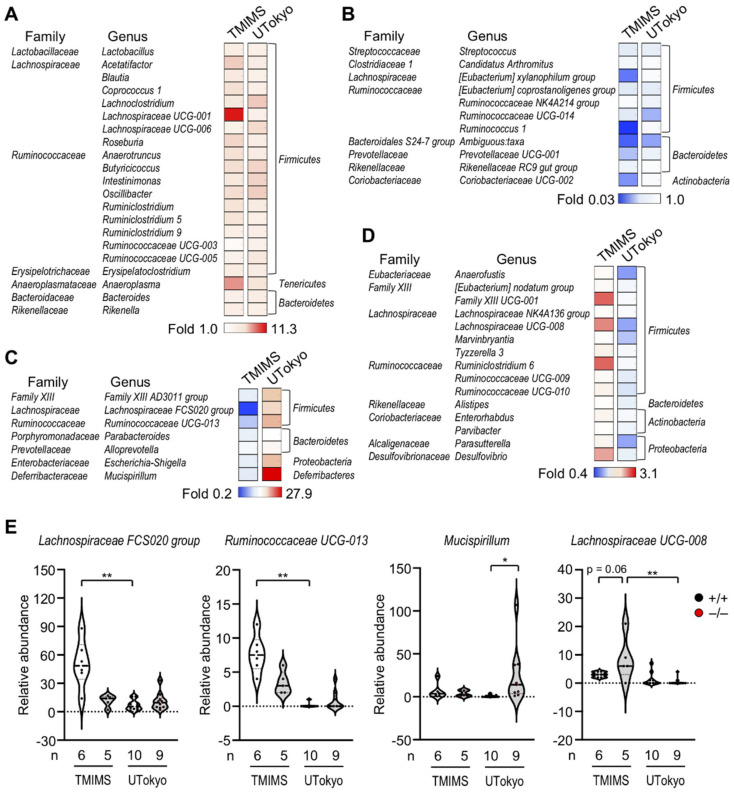
Figure 4.
The effects of sPLA2-IIA deficiency on the gut microbiota in the TMIMS and UTokyo animal facilities. Metagenome analysis of bacterial 16S RNA in the stool was performed as described previously [48]: (A,B) Heatmap representation of bacterial genera commonly increased (A) or decreased (B) in Pla2g2a−/− mice relative to Pla2g2a+/+ mice in both facilities. (C,D) Heatmap representation of bacterial genera that were increased in the UTokyo facility and decreased in the TMIMS facility (C) and vice versa (D) in Pla2g2a−/− mice relative to Pla2g2a+/+ mice. (E) Violin plots of the abundance of representative bacteria in Pla2g2a+/+ and Pla2g2a−/− mice. Values are mean ± SEM. *, p < 0.05; **, p < 0.01. Statistical analysis was performed with one-way ANOVA followed by Šidák multiple comparison test, or Kruskal–Wallis test followed by Dunn’s multiple comparison test (E). The numbers of mice used for the analysis are indicated in each panel.