Figure 1.
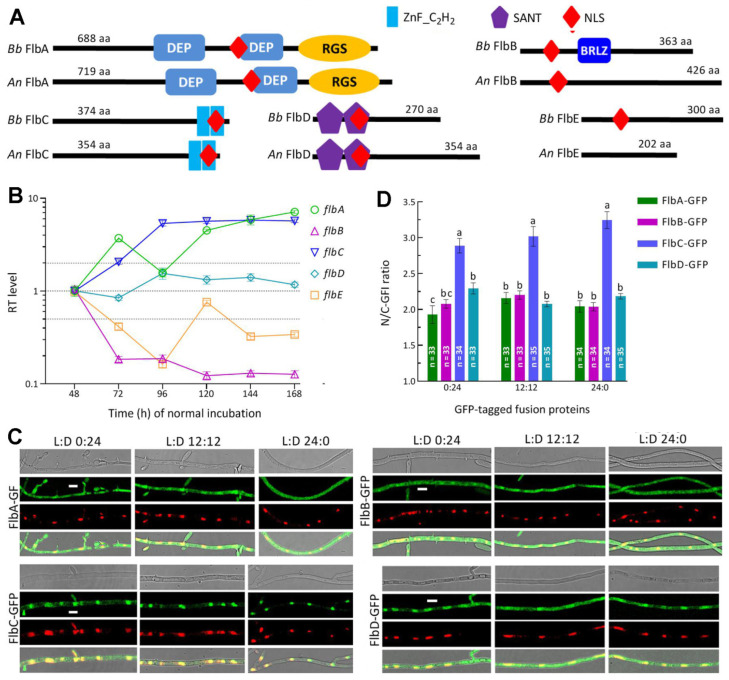
Sequence, transcription and subcellular features of FlbA–FlbE in B. bassiana. (A) Sequence comparison of FlbA–FlbE between B. bassiana (Bb) and A. nidulans (An). Domains and nuclear localization signal (NLS) were predicted from each protein sequence at http://smart.embl-heidelberg.de/ (accessed on 20 March 2022) and http://nls-mapper.iab.keio.ac.jp/ (accessed on 20 March 2022), respectively. (B) Relative transcript (RT) levels of flbA–flbE in the SDAY cultures of wild-type Bb strain (WT) during a 7-d incubation at the optimal regime of 25 °C in a light/dark (L:D) cycle of 12:12 with respect to the standard level on day 2. (C) LSCM images (scales: 5 μm) for subcellular localization of green fluorescence-tagged Flb fusion proteins expressed in the WT strain. Cell samples were taken from the 3 d-old SDBY cultures grown at 25 °C in the respective L:D cycles of 0:24, 12:12 and 24:0 and stained with the nuclear dye DAPI (shown in red). Each four-image panel show bright, expressed (green), stained (red nuclei) and overlapped (yellow nuclei) images of the same field, respectively. (D) Nuclear versus cytoplasmic green fluorescence intensity (N/C-GFI) ratios of the fusion proteins in the hyphal cells. Error bars denote standard deviations (SDs) of the means from three cDNA samples analyzed via qPCR (B) or 33 to 35 cells in the examined hyphae (D).