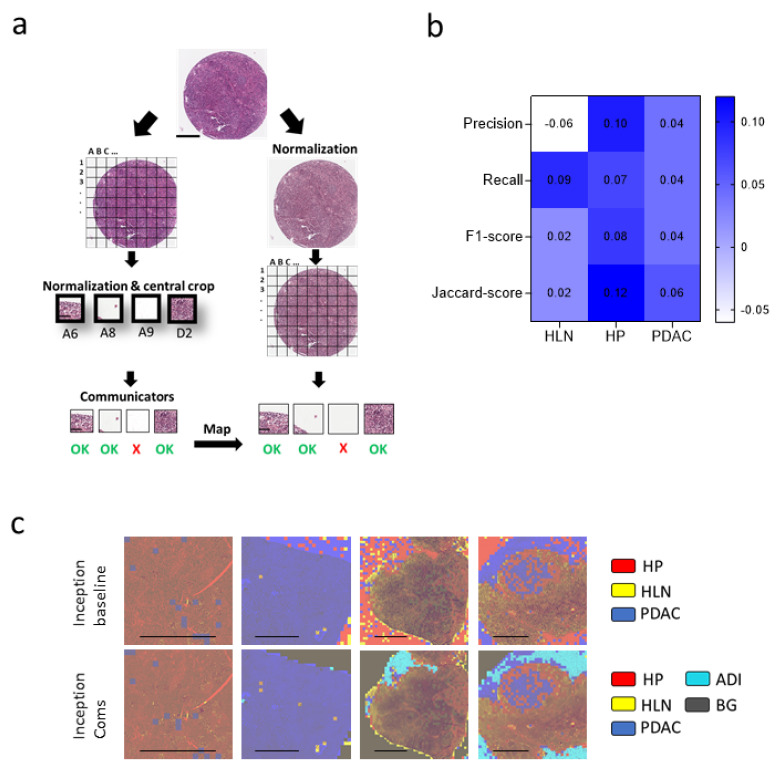
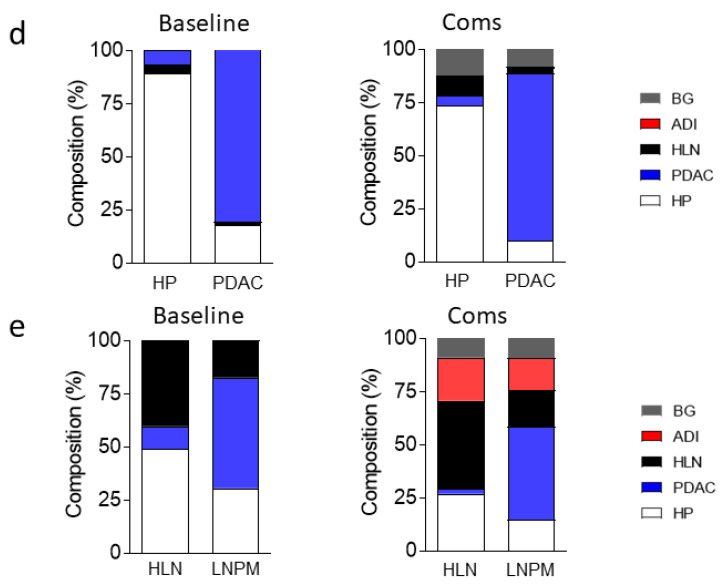
Figure 7.
Communicator-based clean-up can be transferred to CNNs using a different input size. (a) Selection of the normalized 299 × 299 × 3 tissue patches based on the classification of the communicator CNNs using a 224 × 224 × 3 crop of the image tiles is illustrated (Spot: Scale bar = 300 μm, Patch: Scale bar = 60 μm). (b) Heatmap of the performance difference between the inceptionnetv3 trained on data with and without the data clean-up via the communicators is shown. (c) Visualization of the classified validation data by the baseline or cleaned-up inceptionv3 net. Healthy lymph node (HLN, yellow), healthy pancreas (HP, red), pancreatic ductal adenocarcinoma (PDAC, blue), background (BG, grey) and adipose tissue (ADI, cyan) classification is illustrated (scale bar = 2 mm). (d) Pooled classification, as determined using a baseline and cleaned inceptionv3 network from healthy pancreas (HP) (n = 3) and pancreatic ductal adenocarcinoma (PDAC) images (n = 15), is illustrated. (e) Average of classification, as determined using a baseline and cleaned inceptionv3 network of images showing healthy lymph nodes (HLN) (n = 5) and lymph nodes with pancreatic ductal adenocarcinoma metastasis (LNPM) (n = 6), is presented (ADI= adipose tissue, BG = background).