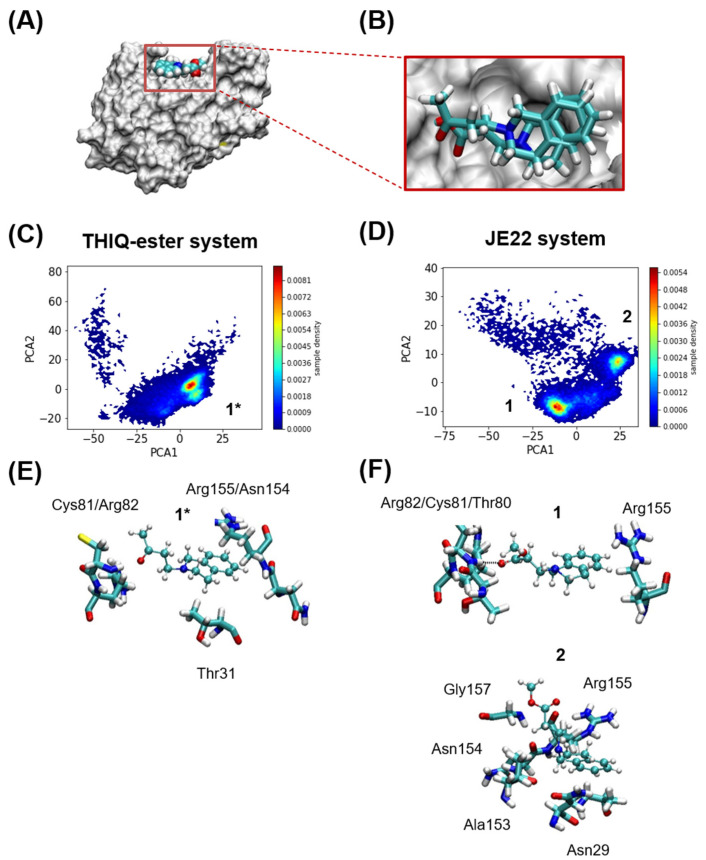
Figure 2.
Computational analysis. (A) Murine CD44 HABD in MSMS representation in white, and THIQ-Ester in VDW representation at the position where it is found in the crystal structure PDB id 5BZK, used as starting point of the MD simulations. (B) Relative positions of JE22 and THIQ-ester ligands at the start of the MD simulations. The JE22 molecule was aligned using the position of the THIQ-ester ligand in the crystal structure PDB id 5BZK. (C,D) Comparison of MD-resolved ligand binding poses (1* for THIQ-ester system and 1 and 2 for the JE22 system). Probability density of the first two principal of the multidimensional matrix built with the distances between either the nitrogen and oxygen atoms of (C) THIQ-ester or (D) JE22, and any of the nitrogen or oxygen atoms of the residues that define the THIQ-ester binding pocket. The binding pocket where THIQ-ester is found crystallographically is formed by residues Asn29, Thr31, Glu41, Thr80, Cys81, Arg82, and Arg155; any of these residues have at least one atom at 3 Å of the ligand. (E,F) Representative structures of each cluster are shown in panels (E,F) for THIQ-ester and JE22 systems, respectively. The residues from the protein where there is at least an atom within 2.5 Å of the ligand are shown in licorice representation.