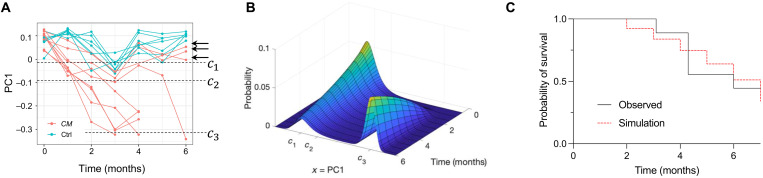
Fig. 4. Validation of the AML state-space and state-transition model.
(A) Using only the state-space PC loadings , samples from an independent cohort of CM and control mice were projected into the state-space (CM, n = 9; control, n = 7). Control and CM state-transition trajectories were accurately identified, including three CM mice that did not develop leukemia at the experiment end point (black arrows). (B) The solution of the FP model of state-transition gives the evolution of the probability of the miRNA transcriptome particle being at a point in the state-space at any given time. (C) The solution of the FP equation with parameters derived from the training cohort is integrated over the state-space past the transition point c2 to calculate the probability to develop AML over time. Using a Gaussian kernel with the initial state of the miRNA transcriptome following CM induction [t = 1 shown in (A)], the FP equation accurately predicts AML in the validation cohort of CM mice, including the prediction that not all mice would manifest AML in the 6-month duration of the experiment. The log-rank test was unable to reject the null hypothesis that there is no difference between the observed and simulated curves [P = 0.79, HR = 0.86 (0.26 to 2.8)].