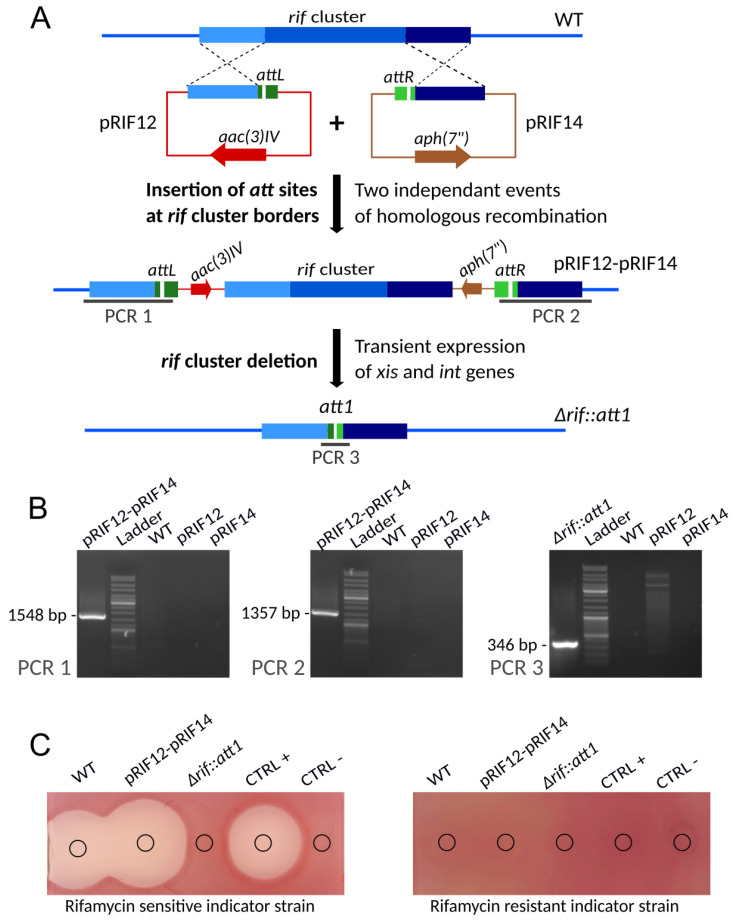
Figure 4.
Construction of an unmarked rif cluster deletion mutant. (A) Schematic representation (not to scale) of the successive steps. First the attL and attR sequences are successively integrated upstream and downstream of the rif cluster via two single homologous recombination events. Then the complete region between the attL and attR sites is excised following the introduction of pEA01, resulting in an unmarked rif cluster deletion mutant containing a 33-bp scar (att1). (B) PCR verification of the strains at different stages of the construction. Verifications are based on three PCRs. The extent of the expected amplicons for each of the PCR is indicated in panel A. Lanes: pRIF12-pRIF14-genomic DNA of A. mediterranei DSM43770 harboring pRIF12 and pRIF14, representative of all analyzed exconjugants; Ladder-GeneRuler DNA Ladder (SM0331 Thermo Scientific); WT-genomic DNA of A. mediterranei DSM43770 wild-type strain; pRIF12-Plasmid pRIF12; pRIF14-Plasmid pRIF14; ∆rif::att1-genomic DNA of A. mediterranei with a deletion of the rif cluster. (C) Bioassays. The antibacterial activity of culture supernatants from the wild-type strain A. mediterranei DSM43770, the strain harbouring pRIF12 and pRIF14, and the mutant strain ∆rif::att1 was assayed against S. aureus HG003 (sensitive to rifamycin) and its rpoBH481Y derivative (resistant to rifamycin). The slight difference in inhibition zone size observed for WT and pRIF12-pRIF14 strains is due to the fluctuation of rifamycin production between different flasks. MP5 with or without 5 µg/mL of rifamycin SV was used as positive or negative control, respectively.