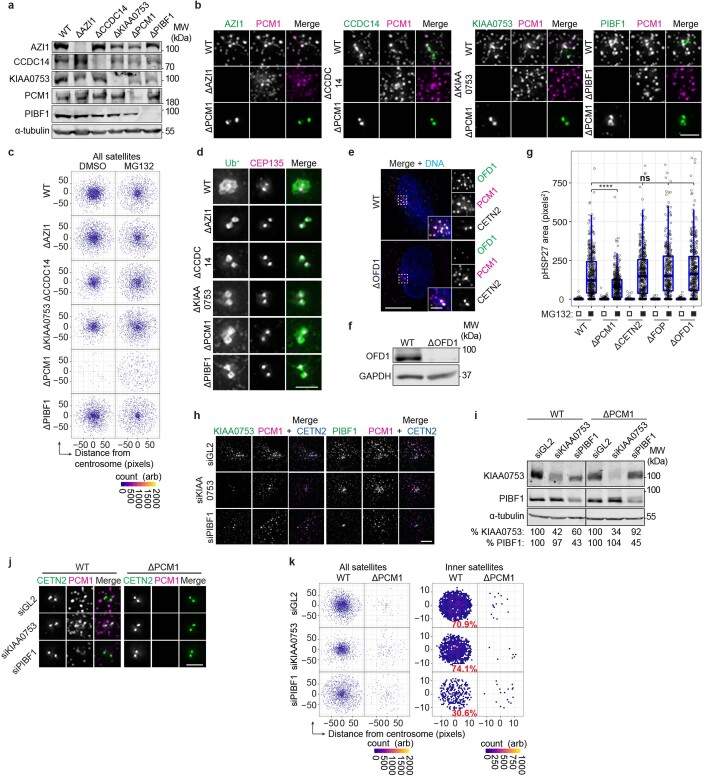
Extended Data Fig. 3. Centriolar satellites are required for aggresome formation.
(a) CRISPR/Cas9 mediated gene disruption was used to generate AZI1, CCDC14, KIAA0753, PCM1 and PIBF1 knockout (KO) RPE-1 cell lines. Extracts from WT and KO cells were probed as indicated. (b) Loss of protein in AZI1, CCDC14, KIAA0753, PCM1 and PIBF1 KO cells was also confirmed by IF microscopy. Scale bar 2 μm. (c) Intensity maps of PCM1 distribution relative to the centrosome in WT and KO cells treated as indicated for 5 hours. Arb, arbitrary units. (d) Ub+ and CEP135 staining in WT and satellite protein KO cells treated with MG132. Scale bar 2 μm. (e) WT and OFD1 KO cells were stained for OFD1, PCM1 and CETN2. Scale bar 10 μm, inset 2 μm. (f) Immunoblot of extracts from WT and OFD1 KO cells probed as indicated. (g) Box-and-whisker plot showing the area occupied by pHSP27 in the indicated cell lines treated with MG132. Boxes represent the median, upper and lower quartiles, whiskers represent 1.5x the interquartile range, with individual values superimposed. n = WT: 400 (DMSO), 414 (MG132); ΔCETN2: 411 (DMSO), 434 (MG132); ΔFOP: 456 (DMSO), 352 (MG132); ΔOFD1: 422 (DMSO), 377 (MG132); ΔPCM1: 519 (DMSO) and 429 (MG132) aggresomes examined over 2 independent experiments. Data were compared using a Kruskal-Wallis ANOVA test and a post-hoc Dunn multiple comparison test performed to calculate p-values. ****p < 0.0001; n.s. not significant. (h) WT cells were transfected with siRNAs and stained as indicated. Scale bar 2 μm. (i) Immunoblot of extracts from WT and PCM1 KO cell lines transfected with siRNAs. All samples were run on the same gels; however, an intervening lane between the WT and PCM1 KO cell lines was excised, thereby placing non-adjacent lanes next to each other in the panel. Numbers at the bottom indicate the mean band intensity expressed as a percentage of signal of the corresponding control for the indicated proteins. (j) WT and PCM1 KO cells treated with control, KIAA0753 or PIBF1 siRNAs were stained as indicated. Scale bar 2 μm. (k) Intensity maps of PCM1 distribution relative to the centrosome in WT and PCM1 KO cells treated and stained as in j. Arb, arbitrary units. Unprocessed immunoblots, numerical data and p values are provided as source data.