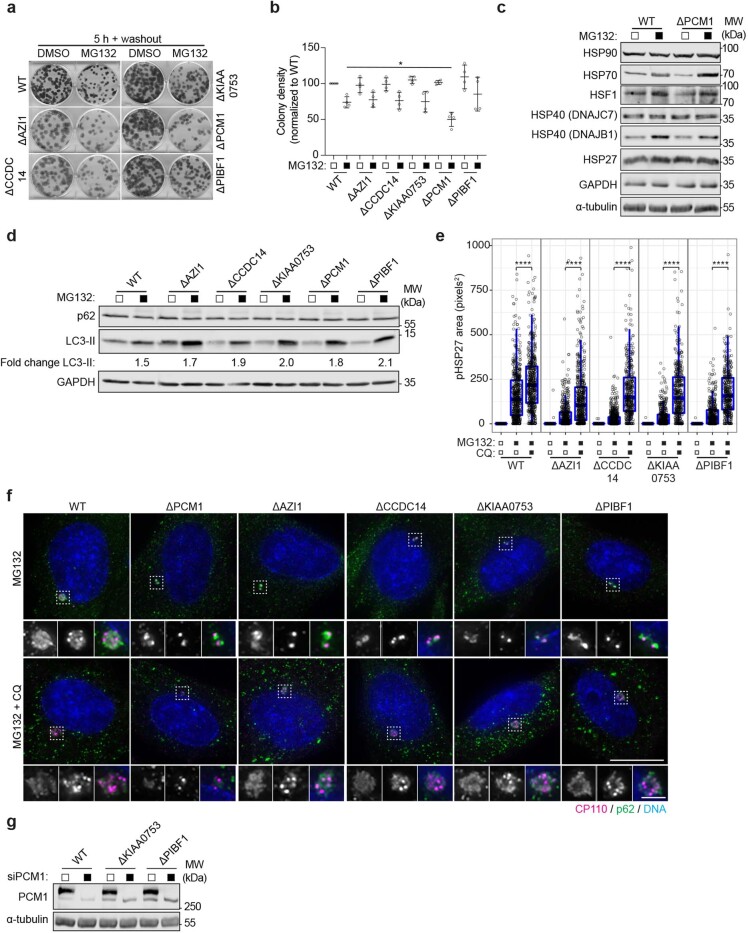
Extended Data Fig. 4. Centriolar satellites direct protein aggregates to the aggresome in the absence of autophagy.
(a) WT, AZI1, CCDC14, KIAA0753, PCM1 and PIBF1 KO cells were seeded for clonogenic assays and treated with MG132 for 5 hours. The drug was then extensively washed out and the cells left to grow for 12 days. (b) Colony density was quantified and growth normalized to WT. Mean ± s.d. is shown, n = 4 from two independent experiments. *p < 0.05 by two-tailed unpaired Student’s t-test. (c) Immunoblots of extracts from WT and PCM1 KO cells treated with DMSO or MG132 probed as indicated. α-tubulin was used as a loading control. (d) Immunoblots of extracts from WT, AZI1, CCDC14, KIAA0753, PCM1 and PIBF1 KO cells treated with DMSO or MG132 and probed as indicated. The fold change in the amount of LC3-II present in MG132 treated cells as compared to the DMSO treated counterpart is indicated. GAPDH was used as a loading control. (e) Box-and-whisker plot showing the area occupied by pHSP27 in WT, AZI1, CCDC14, KIAA0753 and PIBF1 KO cells treated with DMSO, MG132 or MG132 plus chloroquine (CQ). Boxes represent the median, upper and lower quartiles, whiskers represent 1.5x the interquartile range, with individual values superimposed. n = WT: 470 (DMSO), 529 (MG132), 513 (MG132/CQ); ΔAZI1: 484 (DMSO), 458 (MG132), 524 (MG132/CQ); ΔCCDC14: 387 (DMSO), 739 (MG132), 404 (MG132/CQ); ΔKIAA0753: 381 (DMSO), 426 (MG132), 411 (MG132/CQ); ΔPIBF1: 470 (DMSO), 529 (MG132) and 513 (MG132/CQ) aggresomes examined over 2 independent experiments. Data were compared using a Kruskal-Wallis ANOVA test and a post-hoc Dunn multiple comparison test performed to calculate p-values. ****p < 0.0001. (f) WT, PCM1, AZI1, CCDC14, KIAA0753 and PIBF1 cells were treated with MG132 or MG132 plus CQ and stained for CP110 and p62. Scale bar 10 μm, 2 μm. (g) Immunoblot of extracts from WT, KIAA0753 and PIBF1 KO cells transfected with siRNAs against PCM1. α-tubulin was used as a loading control. Unprocessed immunoblots, numerical data and p values are provided as source data.