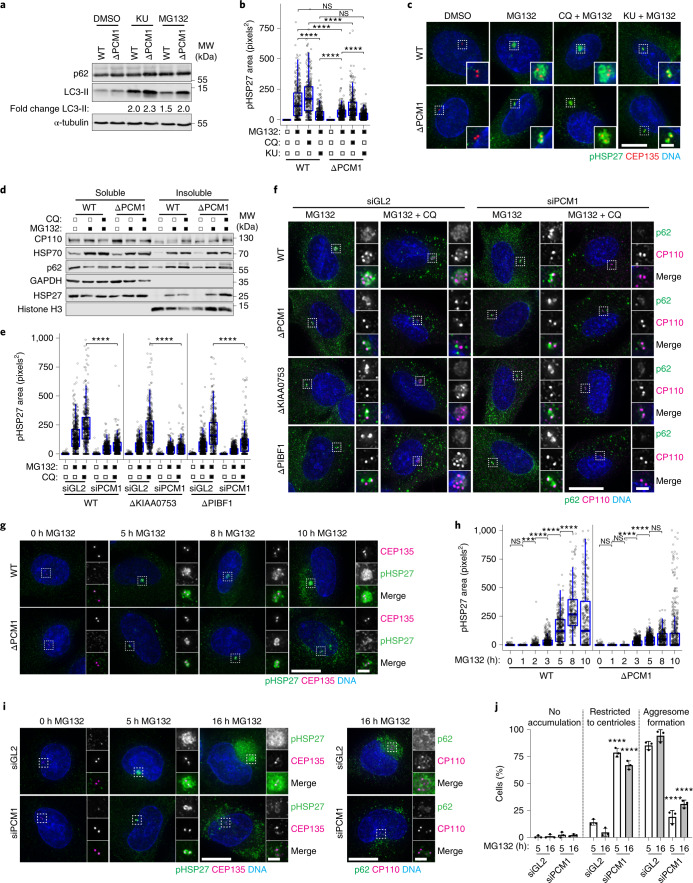
Fig. 4. Centriolar satellites direct protein aggregates to the aggresome in the absence of autophagy.
a, WT and ΔPCM1 cells treated and probed as indicated. KU, KU-0063794. b, pHSP27 area in WT and ΔPCM1 cells treated as indicated. n = 360 (WT-DMSO), 368 (WT-MG132), 323 (WT-MG132 + CQ), 386 (WT-MG132 + KU), 360 (ΔPCM1-DMSO), 375 (ΔPCM1-MG132), 366 (ΔPCM1-MG132 + CQ) and 418 (ΔPCM1-MG132 + KU) aggresomes examined over 2 independent experiments. ****P < 0.0001. c, Cells treated as in b were stained as indicated. d, Fractions from WT and ΔPCM1 cells treated and probed as indicated. e, pHSP27 area in siRNA-transfected WT, ΔKIAA0753 and ΔPIBF1 cells treated as indicated. Number of aggresomes examined over 2 experiments: WT: n = 522 (siGL2-DMSO), 525 (siGL2-MG132), 433 (siGL2-MG132 + CQ), 466 (siPCM1-DMSO), 422 (siPCM1-MG132) and 460 (siPCM1-MG132 + CQ); ΔKIAA0753: n = 509 (siGL2-DMSO), 563 (siGL2-MG132), 503 (siGL2-MG132 + CQ), 474 (siPCM1-DMSO), 442 (siPCM1-MG132) and 380 (siPCM1-MG132 + CQ); ΔPIBF1: n = 680 (siGL2-DMSO), 608 (siGL2-MG132), 478 (siGL2-MG132/CQ), 497 (siPCM1-DMSO), 531 (siPCM1-MG132) and 427 (siPCM1-MG132/CQ). ****P < 0.0001. f, siRNA-transfected WT cells and ΔPCM1, ΔKIAA0753 and ΔPIBF1 cells were treated and stained as indicated. g, WT and ΔPCM1 cells were treated and stained as indicated. h, pHSP27 area in WT and ΔPCMI cells treated as indicated. Number of aggresomes examined over 2 independent experiments: WT: n = 423 (0 h), 387 (1 h), 428 (2 h), 470 (3 h), 470 (4 h), 497 (5 h), 392 (8 h) and 287 (10 h); ΔPCM1: n = 524 (0 h), 581 (1 h), 580 (2 h), 598 (3 h), 640 (4 h), 635 (5 h), 593 (8 h) and 394 (10 h). ****P < 0.0001, **P < 0.01. i, siRNA-transfected A-375 cells were treated and stained as indicated. j, Quantitation of aggresome formation in A-375 cells transfected with control or PCM1 siRNAs, then treated with MG132 as indicated. Data displayed as the mean ± s.d., n = 3 independent experiments. ****P < 0.0001 by two-tailed unpaired Student’s t-test. For b, e and h, boxes represent the median, upper and lower quartiles, whiskers represent 1.5× the interquartile range, with individual values superimposed. Data compared using Kruskal–Wallis ANOVA test and post-hoc Dunn multiple comparison test. Scale bars, 10 μm (c,f,g,i) or 2 μm (insets of c,f,g,i). Unprocessed immunoblots, numerical data and P values are provided as source data.