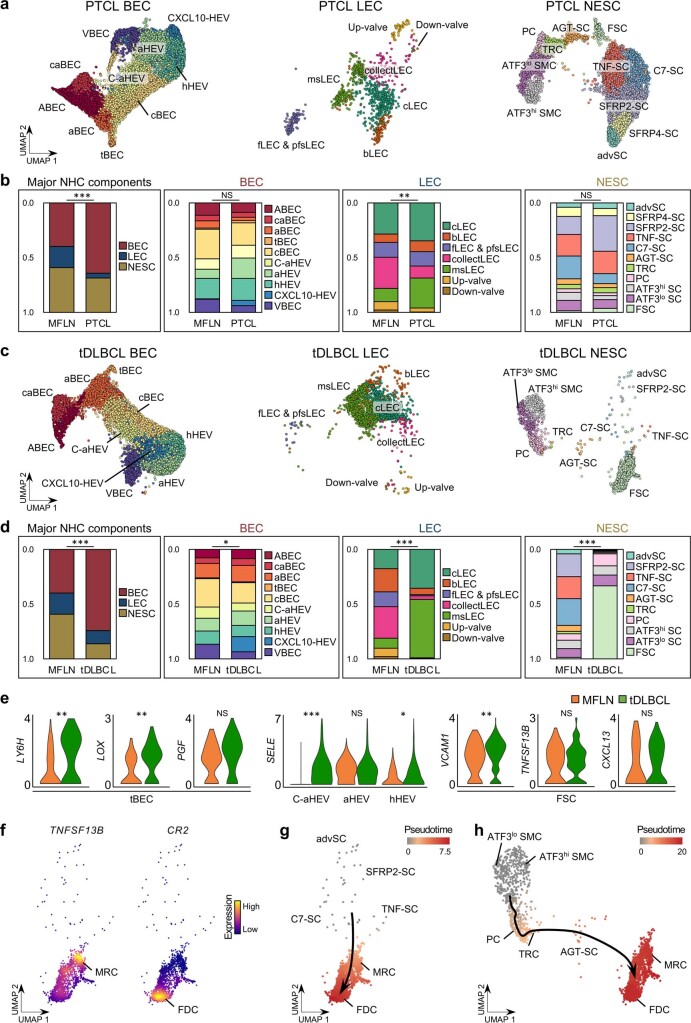
Extended Data Fig. 10. Applicability of the LNNHC atlas to PTCL and tDLBCL stroma.
a, UMAP plots of PTCL BEC (left), LEC (middle), and NESC (right) subclusters. b, Compositional differences between PTCL and MFLN NHCs based on major NHC components, BEC subclusters, LEC subclusters, and NESC subclusters (from left to right). **P = 0.0076, ***P = 2.7 × 10−4 (two-sided chi-squared test). NS, not significant. c, UMAP plots of tDLBCL BEC (left), LEC (middle), and NESC (right) subclusters. d, Compositional differences between tDLBCL and MFLN NHCs based on major NHC components, BEC subclusters, LEC subclusters, and NESC subclusters (from left to right). *P = 0.030, ***P = 3.4 × 10−6 (Major NHC components), ***P = 6.3 × 10−16 (LEC), ***P = 1.1 × 10−23 (NESC) (two-sided chi-squared test). e, Violin plots comparing expressions of key genes between MFLN (orange) and tDLBCL (green) samples according to selected NHC subclusters. *P = 0.015, **P = 0.0039 (LY6H), **P = 0.0090 (LOX), **P = 0.0075 (VCAM1), ***P = 8.0 × 10−111 (two-sided Wilcoxon Rank-Sum test with Bonferroni correction). NS, not significant. f, Expression of TNFSF13B (left) and CR2 (right) in tDLBCL follicular stromal cells identifying MRCs and FDCs, respectively. g,h, Pseudo-time developmental stages in tDLBCL advSCs, SFRP2-SCs, TNF-SCs, C7-SCs, MRCs, and FDCs (g) or in tDLBCL SMC subclusters, PCs, TRCs, AGT-SCs, MRCs, and FDCs (h). Dark winding lines in the cell objects indicate putative developmental trajectories. The statistical source data are provided.