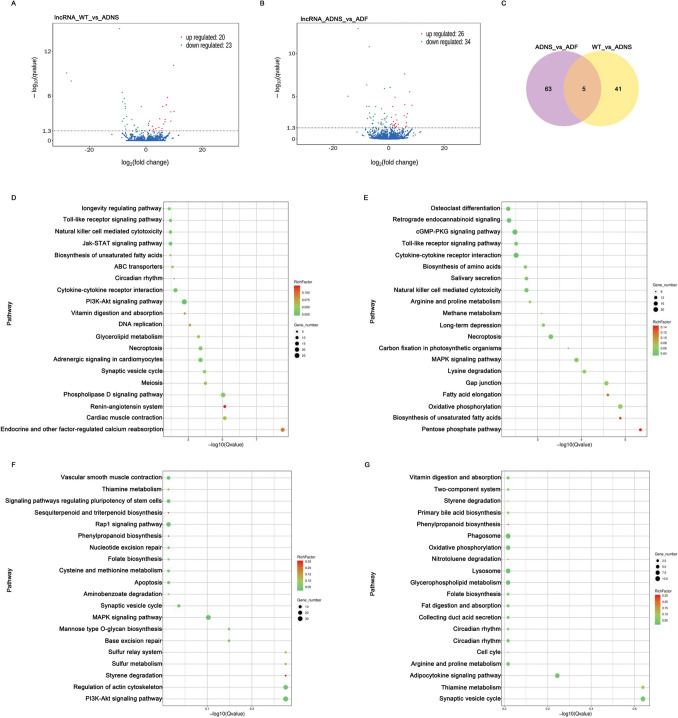
Figure 3.
Differentially expressed lncRNAs in hippocampal tissues from mice in each group. (A) The number of upregulated (22) and downregulated (24) lncRNAs between ADNS and WT are shown. (B) The number of upregulated (29) and downregulated (39) differentially expressed lncRNAs between ADF and ADNS are shown. (C) Venn showed 5 lncRNAs at the intersection of ADNS versus ADF and WT versus ADNS. (D) KEGG analysis involving the top 20 terms of differentially expressed lncRNA co-located with mRNA in WT versus ADNS. Most genes focus in PI3K-AKT signal pathway. (E) KEGG analysis involving the top 20 terms of differentially expressed lncRNA co-located with mRNA in ADNS versus ADF. Most genes focus in cytokine-cytokine receptor interaction and cGMP-PKG signaling pathway. (F) KEGG analysis of differentially expressed lncRNA co-expressed with mRNA in WT versus ADNS. Most genes focus on the pathway of PI3K-Akt, MAPK, regulation of actin cytoskeleton and Rap1 signaling pathway. (G) KEGG analysis of differentially expressed lncRNA co-expressed with mRNA in ADNS versus ADF. Most genes focus on the pathway of phagosome, synaptic vesicle cycle and adipocytokine signaling pathway (KEGG pathways: https://www.kegg.jp/kegg/kegg1.html).