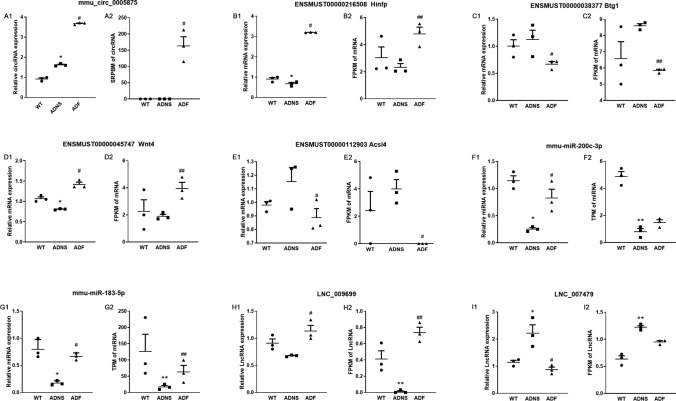
Figure 6.
Validation of Differentially Expressed RNAs in hippocampal tissues from mice in each group. (A) Expression changes for select circRNA using RT-PCR (left) and high-throughput sequencing results (right) among WT, ADNS, and ADF samples. (B–E) Expression changes for select mRNAs using RT-PCR (left) and high-throughput sequencing results (right) among WT, ADNS, and ADF samples. (F,G) Expression changes for select miRNA using RT-PCR (left) and high-throughput sequencing results (right) among WT, ADNS, and ADF samples. (H,I) Expression changes for select lncRNAs using RT-PCR (left) and high-throughput sequencing results (right) among WT, ADNS, and ADF samples. *p < 0.05, **p < 0.01 versus WT; #p < 0.05, ##p < 0.01 versus ADNS. The Dunnett’s test were used for statistical analysis.