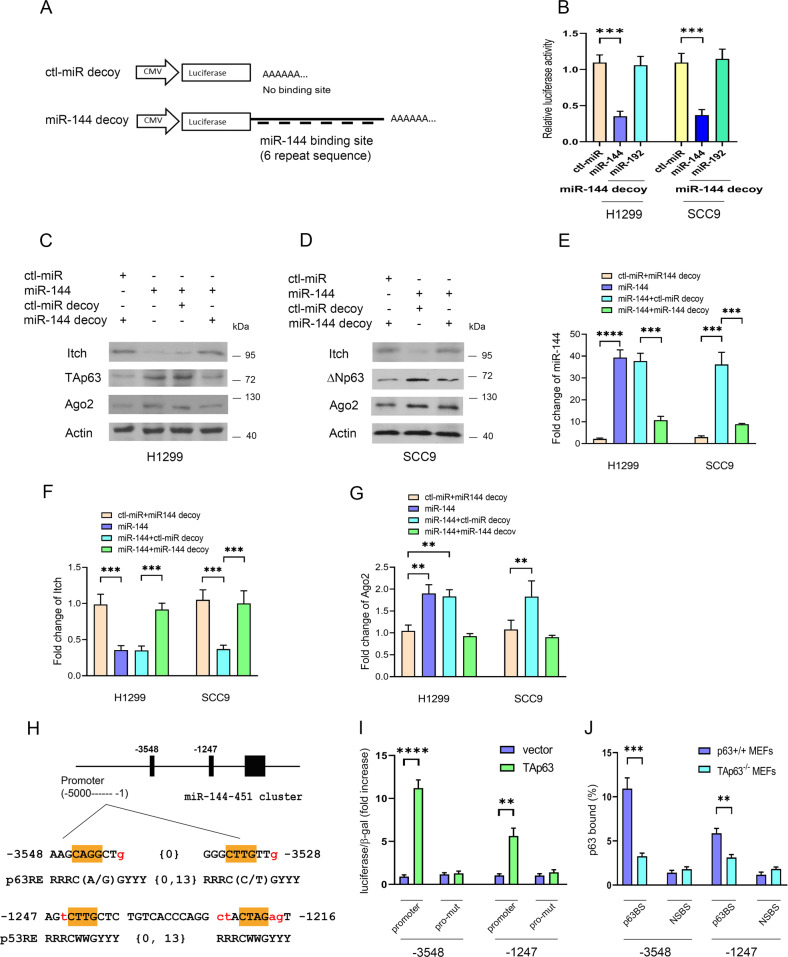
Fig. 6. miR-144 regulates p63.
A A miR-144 decoy was generated in which six tandem miR-144 sequences complementary to miR-144 were linked to a luciferase reporter gene. B Plasmids expressing control-miRNA, miR-144, miR-192, or in combination with miR-144 decoy were transfected into H1299 and SCC9 cells. Luciferase activities were measured. C Plasmids expressing control-miRNA, miR-144, control-miRNA decoy, and miR-144 decoy were transfected into H1299 cells. The levels of endogenous Itch, TAp63, and Ago2 proteins were detected by Western blotting as indicated. D Similar to (C), except that SCC9 cells were used. E The expression level of miR-144 was detected by the TaqMan® method. F The level of Itch mRNA was detected by qRT–PCR. G The level of Ago2 mRNA was detected by qRT–PCR. H The two potential p63 binding sites in the promoter of the miR-144/451 cluster were compared to the consensus p63/p53 binding sites as indicated. I The relative luciferase activity in the presence of TAp63 or an empty vector is presented. The reporter containing wt or mutant sequences was examined in H1299 cells. J TAp63 ChIP assays, followed by qRT–PCR for the miR-144/451 cluster promoter using TAp63+/+ MEFs and TAp63−/− MEFs, were performed. p63 binding was measured by qRT–PCR. TAp63BS: TAp63-binding site. NSBS: nonspecific binding site on the promoter of the miR-144/451 cluster. All experiments were performed in triplicate. **p < 0.01; ***p < 0.001; ****p < 0.0001.