The computational values using molecular docking and FPL simulations.
| No. | Compound | ΔGDock | F Max | W | ΔGPreFPLa | Predicted ICPre50 rangeb |
|---|---|---|---|---|---|---|
| 1 | M15 | −9.4 | 692.8 ± 26.7 | 77.7 ± 3.7 | −9.87 | High-nanomolar |
| 2 | M8 | −9.7 | 636.4 ± 37.9 | 77.0 ± 3.1 | −9.82 | High-nanomolar |
| 3 | M11 | −9.6 | 764.9 ± 32.0 | 73.4 ± 3.6 | −9.62 | High-nanomolar |
| 4 | M9 | −9.7 | 601.5 ± 30.2 | 68.5 ± 2.7 | −9.35 | High-nanomolar |
| 5 | M2 | −10.2 | 579.2 ± 49.4 | 60.5 ± 4.6 | −8.90 | Sub-micromolar |
| 6 | M13 | −9.5 | 587.9 ± 37.6 | 59.4 ± 5.3 | −8.84 | Sub-micromolar |
| 7 | M3 | −9.9 | 593.1 ± 35.3 | 59.0 ± 2.5 | −8.82 | Sub-micromolar |
| 8 | M5 | −9.8 | 595.8 ± 30.1 | 58.4 ± 2.1 | −8.78 | Sub-micromolar |
| 9 | M4 | −9.9 | 549.1 ± 32.3 | 56.6 ± 3.9 | −8.68 | Sub-micromolar |
| 10 | M16 | −9.4 | 587.5 ± 37.4 | 54.6 ± 3.8 | −8.57 | Sub-micromolar |
| 11 | M6 | −9.8 | 551.1 ± 24.7 | 54.0 ± 3.0 | −8.54 | Sub-micromolar |
| 12 | M1 | −10.6 | 530.2 ± 20.0 | 50.8 ± 2.4 | −8.36 | Micromolar |
| 13 | M12 | −9.6 | 512.9 ± 28.0 | 44.1 ± 1.3 | −7.98 | Micromolar |
| 14 | M14 | −9.4 | 447.9 ± 19.0 | 40.7 ± 2.5 | −7.79 | Micromolar |
| 15 | M7 | −9.7 | 418.8 ± 30.2 | 37.7 ± 2.9 | −7.62 | Micromolar |
| 16 | M10 | −9.6 | 428.9 ± 20.3 | 34.3 ± 1.8 | −7.43 | Micromolar |
The predicted binding affinity ΔGPreFPL = −0.056 × W − 5.512.59
The predicted ICPre50 was calculated via formula 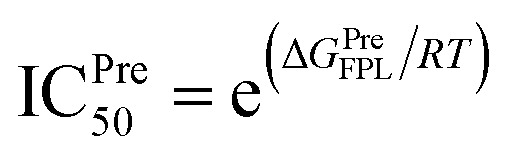 with assumption that IC50 equals to inhibition constant ki. The computed error is the standard error of the mean. The unit of force and energy in pN and kcal mol−1, respectively.
with assumption that IC50 equals to inhibition constant ki. The computed error is the standard error of the mean. The unit of force and energy in pN and kcal mol−1, respectively.
