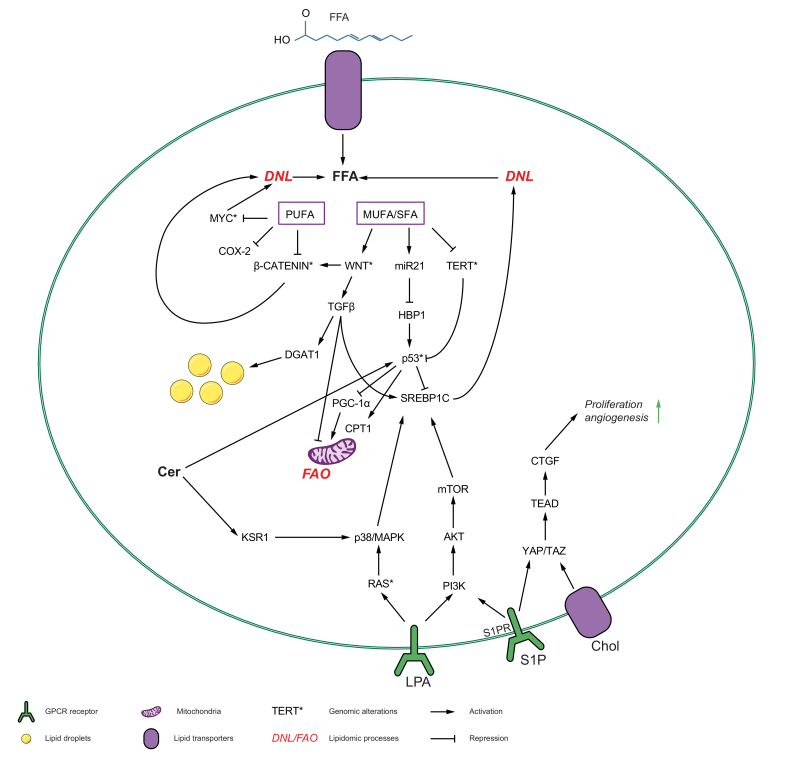
Fig. 2.
The interplay between lipid metabolism and oncogene pathways that leads to tumorigenesis in the liver.
Various lipids influence and are influenced by the recurrently deregulated oncogenic pathways, p53, RAS/MAPK, PI3K/AKT/mTOR signalling axis, Wnt/β-catenin signalling axis, TGF-β signalling axis and myc and TAZ/YAP pathways to cause hepatocarcinogenesis. The inhibited genes are marked with (⊥) and activated with (↓). The recurrent molecular alterations in oncogenes are marked with (∗). CL, cardiolipin; CTGF, connective tissue growth factor; DNL, de novo lipogenesis; FA, fatty acid; GlcCer, glucosylceramide; mTOR, mammalian target of rapamycin; NPC1, NPC intracellular cholesterol transporter 1; PI3K, phosphoinositide 3-kinase; PDK1, phosphoinositide-dependent protein kinase 1; S1P, sphingosine-1-phosphate; SL, sphingolipid; TG, triglycerides; TGF-β, transforming growth factor-β; YAP, Yes-associated protein.