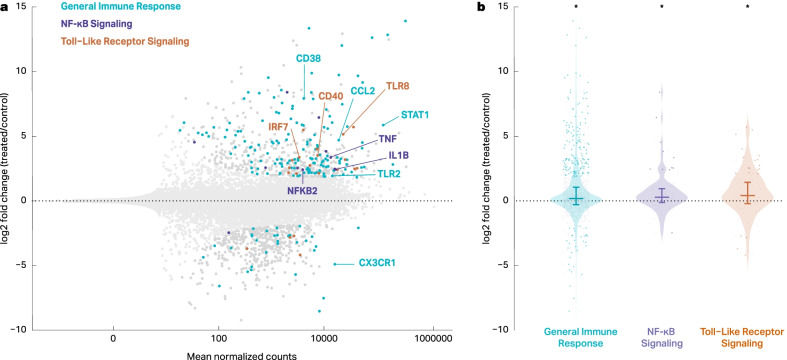
Fig. 4.
LPS/INFγ treatment induces a robust proinflammatory transcriptional change in IMGLs. a Bulk RNASeq was performed in IMGLs treated with 1 µM Aβ oligomers or 10 ng/ml LPS + 20 ng/ml INFγ for 24 h. Differential genes (p < 0.01, absolute fold change > 2) are labeled in dark gray and non-significant genes (p > 0.01, absolute fold change < 2) are labeled in light gray. Blue, purple, and orange represent differential genes associated with general immune response, NF-κB signaling, and toll-like receptor signaling, respectively. b Log2 fold-changes for the genes in specific pathways shown in 4A. Non-significant genes are shown in light gray and differential genes are shown in blue, purple, and orange, respectively, for general immune response, NF-κB signaling, and toll-like receptor signaling. 1 Sample Wilcoxon test showed significant differences for the genes in each pathway vs 0 (p = 6.38 × 10–18, p = 1.28 × 10–4, p = 1.88 × 10–4, respectively)