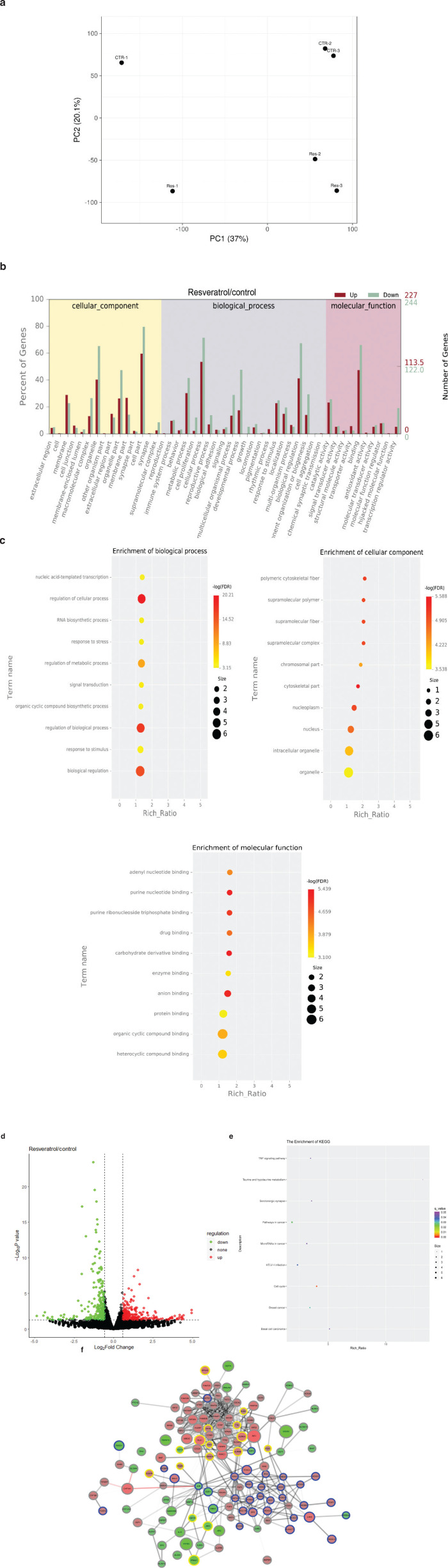
Fig. 3.
RNA sequencing (RNA-seq) reveals the transcriptome in response to resveratrol. (a) PCA-SVD with imputation was used to calculate principal components. CTR: control. Res: resveratrol. (b) Enrichment of the significantly upregulated or downregulated genes at the secondary GO terms. The abscissa is the GO term. The left ordinate is the percentage of the number of genes, and the right ordinate is the number of genes. (c) GO analysis. The abscissa represents the enrichment ratio, and the ordinate represents different GO items. (d) Volcano plot showing genes with significant changes in RNA sequencing. The abscissa is the logarithm of the fold change in differential protein expression in the comparison group. Each point represents a specific gene. Genes that were significantly upregulated are labeled with red, and genes that were significantly downregulated are labeled with green. Genes that were not significantly differentially expressed are labeled with black. (e) KEGG analysis. The abscissa is the enriching ratio, and the ordinate represents the name of the KEGG pathway. The size of the dot represents the number of differential genes in this pathway, and the color represents the degree of enrichment of the KEGG entry. (f) Genes showing dramatic changes were visualized by STRING database. The cutoff was set as 1.5-fold change and P < 0.05. The mRNA level reduced by resveratrol is shown in red, and the mRNA level increased by resveratrol is shown in green. The size of the circle indicates the fold change, and the large circle indicates a higher fold change. Transcriptional regulatory genes are highlighted by blue circles, and the kinases are highlighted by yellow circles.