Abstract
RUVBL1 and RUVBL2 are highly conserved AAA ATPases (ATPases Associated with various cellular Activities) and highly relevant to the progression of cancer, which makes them attractive targets for novel therapeutic anticancer drugs. In this work, docking-based virtual screening was performed to identify compounds with activity against the RUVBL1/2 complex. Seven compounds showed inhibitory activity against the complex in both enzymatic and cellular assays. A series of pyrazolo[1,5-a]pyrimidine-3-carboxamide analogs were synthesized based on the scaffold of compound 15 with inhibitory activity and good potential for structural manipulation. Analysis of the structure-activity relationship identified the benzyl group on R2 and aromatic ring-substituted piperazinyl on R4 as essential for inhibitory activity against the RUVBL1/2 complex. Of these, compound 18, which has IC50 values of 6.0 ± 0.6 μM and 7.7 ± 0.9 μM against RUVBL1/2 complex and RUVBL1 respectively, showed the most potent inhibition in cell lines A549, H1795, HCT116, and MDA-MB-231 with IC50 values of 15 ± 1.2 μM, 15 ± 1.8 μM, 11 ± 1.0 μM, and 8.9 ± 0.9 μM respectively. A docking study of the compound was performed to predict the binding mode of pyrazolo[1,5-a]pyrimidine-3-carboxamides. Furthermore, mass spectrometry-based proteomic analysis was employed to explore cellular proteins dysregulated by treatment with compounds 16, 18, and 19. Together, the data from these analyses suggest that that compound 18 could serve as a starting point for structural modifications in order to improve potency, selectivity, and pharmacokinetic parameters of potential therapeutic molecules.
Keywords: RUVBL1; Pontin; RUVBL2; Reptin; AAA ATPase; Organic Synthesis; Docking; pyrazolo[1,5-a] pyrimidine-3-carboxamide; Proteomics
Graphical Abstract
1. Introduction
RUVBL proteins RUVBL1 (TIP49, TIP49a, Rvb1, Pontin) and RUVBL2 (TIP48, TIP49b, Rvb2, Reptin) are present in the Archaea and Eukarya domains, including in humans and fungi. RUVBL proteins are highly conserved AAA proteins (ATPases Associated with various cellular Activities), known to be involved in transcription regulation, DNA damage repair, cell cycle control, and stress adaptation [1-3]. RUVBLs assemble as heteromeric complexes that form an integral component of chromatin remodeling complexes TIP60 [4], INO80 [5] and SWR1/SRCAP [6], as well as of the R2TP chaperone complex [7] and PAQosome [8]. The RuvBL1/2 complex functions as a chaperone itself during the biogenesis of telomerase, H/ACA RNPs and other supramolecular complexes [2, 4, 9, 10]. As well, RuvBL1 and RuvBL2 function independently and even antagonistically in many instances [11-13]. Numerous structural studies indicate that the assembled complex can occur in both hexameric and dodecameric (stacked double hexamer) arrangements [9, 14-17].
There is increasing evidence that the presence of RUVBL1 and RUVBL2 contributes to cancer progression. Overexpression of RUVBL1 and RUVBL2 is observed in many cancer types, including liver, colorectal, breast, gastric, and non-small cell lung cancer [3, 18-22]. Furthermore, multiple studies have suggested that depletion of RUVBLs can hinder cancer cell growth and progression in both in vitro and in vivo models [2, 23, 24]. Combined, this data indicates that RUVBL1 and RUVBL2 may be good targets for anticancer treatment.
Elkaim et al. used virtual screen and enzymatic assays to identify four small-molecule inhibitors of RUVBL1 (Akt1/2 inhibitor, PTP1B inhibitor, Rottlerin, and Pranlukast, see Figure 1) [25], which inhibit RUVBL1 with IC50 values of 24, 15, 10 and 13 μM, respectively. The mechanism by which their action against RUVBL1 occurs needs to be determined in further study. Encouraged by previously published data from RUVBL1 inhibitors, Elkaim et al. focused on two scaffolds, 4-hydroxy-2-pyridone and 4-hydroxy-2-quinolone, and synthesized derivatives; compounds 5 and 6 (Figure 1) inhibited RUVBL1 activity in an ATP-dependent manner with IC50 values of 9 and 18 μM, respectively. Compound 5 demonstrated antiproliferative activity against KB and HL60 cell lines with IC50s of 9 and 15 μM, manifested as apoptotic cell death together with necrosis [26].
Figure 1.
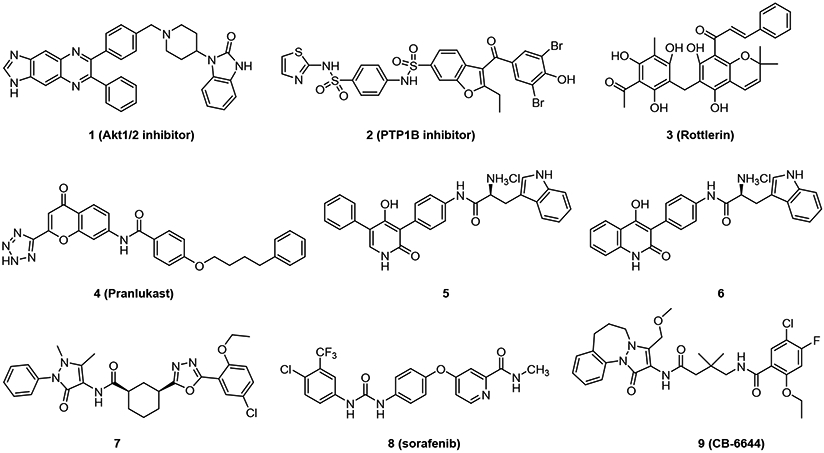
Chemical Structures of known inhibitors against RUVBL proteins
Compound 7 (Figure 1) was initially identified as an RUVBL1/2 complex inhibitor with an IC50 of 59 nM [27]. In cellular assays, compound 7 depleted PIKK-family members DNA-PKcs (PRKDC), ATM, and ATP, all of which require RUVBL1/2 ATPase activity for stability [28]. Compound 7 was shown to arrest NSCLC cells in S phase, which eventually underwent apoptosis, indicated by the sub-G1 population in DNA content analysis. Sorafenib (Figure 1, Compound 8) was identified as a RUVBL2 inhibitor with a Kd value of about 22 μM by high-throughput screening [29]. Sorafenib is shown to be an oral multi-kinase inhibitor used to treat advanced renal cell, liver and thyroid cancer [30]. Sorafenib inhibited RUVBL2 in a mixed non-competitive mode and inhibited activity of the RUVBL1/2 complex. Anderson and co-workers identified CB-6644 as a selective, noncompetitive inhibitor of the RUVBL1/2 complex with a IC50 value of 15 nM [31]. However, CB-6644 showed a maximal inhibition of only ~50%. Herein, we report the discovery of five pyrazolo[1,5-a]pyrimidine-3-carboxamides with the inhibition of RUVBL1/2 ATPase activity and the IC50 value of compound 18 is about 6 μM. Mass spectrometry-based proteomic analysis of cells treated with compounds 16, 18, or 19 were completed to compare the cellular pathways affected.
2. Results and Discussion
2.1. Docking-Based virtual Screening
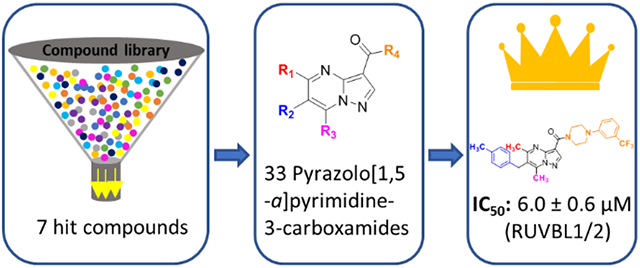
In order to identify novel compounds with anti-RUVBL1/2 activity, docking-based virtual screening was performed using approximately 260,000 compounds from the NCI open compound library. The protein PDB file 5oaf was downloaded from Protein Data Bank (rcsb.org), followed by removal of water molecules and addition of hydrogens in AutoDock 4.2.6 [32]. The coordinates of the Grid center and box size were generated in AutoDock 4.2.6 and the docking results were ranked by binding affinity. Eighty compounds were selected from the top 3000 binding poses after visualization using the BIOVIA Discovery Studio Visualizer 4.5 (Biovia Inc.). In parallel, ligand-based similarity searches in the Swiss Similarity database [33] were performed to screen small molecules from the ChemDiv compound set (around 1,746,000) using CB-6644 as the template molecule. Seventeen compounds (including compounds 16-20) were selected from the top 500 compounds after evaluating their structures.
2.2. Biological evaluation of compounds from virtual screening
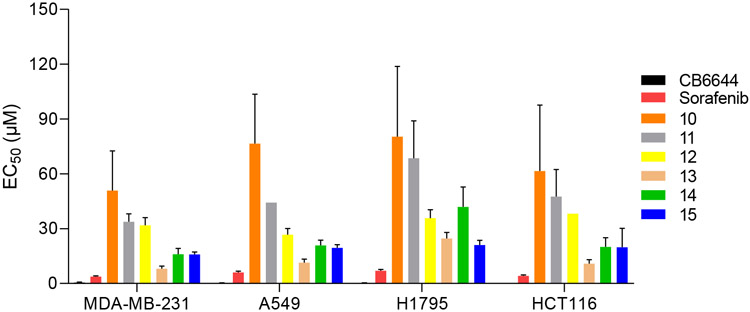
All 97 of the selected compounds were evaluated using an ATPase Assay. Seven of these compounds showed inhibition activity against the RUVBL1/2 complex at an enzymatic level in this work, including compounds 8 and 10-15 (Figure 2, Table 1). Interestingly, after we checked the structures of these compound, we found 8 is known as sorafenib, which was reported as an RUVBL2 inhibitor previously [29]. In addition to activity against the RUVBL1/2 complex, compounds 11, 13, 14, and 15 showed inhibitory activity against RUVBL1. Compound 15 was the only compound to show inhibition towards RUVBL1, RUVBL2, and the RUVBL1/2 complex. Next, we used four cancer cell lines to evaluate anti-proliferative effects of these compounds. However, no compound exhibited more potent inhibition than CB-6644 on any of these cell lines (Figure 3). Compounds 8, 12, 13, 14, and 15 showed moderate inhibitory activity against the RUVBL1/2 complex in enzymatic assays. CB-6644 showed the most potent inhibition in cell lines A549, HCT116, H1795 and MDA-MB-231 (Figure 3). Compared to 14, compound 15 showed comparable inhibition activity in A549, HCT116, and MDA-MB-231 and more potent inhibition in the H1795 cell line.
Figure 2.
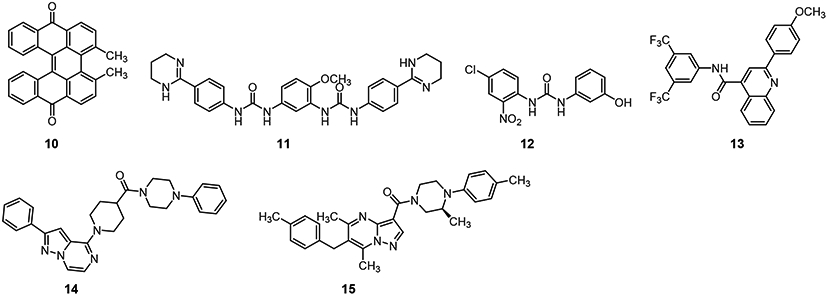
Chemical Structure of compounds active against RUVBL1/2 complex
Table 1.
The inhibitory activity of active compounds from virtual screening
| Compounds | IC50 (μM) |
||
|---|---|---|---|
| RUVBL1/2 | RUVBL1 | RUVBL2 | |
| CB-6644 | 6.1 ± 0.5 | > 66 | > 66 |
| 8 | 55 ± 9.0 | > 66 | > 66 |
| 10 | 77 ± 24 | > 66 | > 66 |
| 11 | 46 ± 1.1 | 39 ± 7.7 | > 66 |
| 12 | 68 ± 11 | > 66 | > 66 |
| 13 | 47± 2.0 | 26 ± 7.2 | > 66 |
| 14 | 35 ± 6.4 | 31 ± 10 | > 66 |
| 15 | 36 ± 2.1 | 65 ± 11 | 29 ± 7.4 |
Figure 3.
Anti-proliferative effects of active molecules from virtual screening. Compounds CB6644, 8 (Sorafenib), 10, 11, 12, 13, 14, and 15 were evaluated for their anti-proliferative effects on breast cancer cell line MDA-MB-231, non-small cell lung cancer cell lines A549 and H1795, as well as human colon cancer cell line HCT116
2.3. Synthesis of pyrazolo[1,5-a]pyrimidine analogs
Due to the in vitro activity and the scaffold that compounds 14 and 15 are sharing, we selected compound 15 as the target compound on which to perform further structural optimization. The pyrazolo[1,5-a]pyrimidine-3-carboxamides were synthesized in a three-step sequence of reactions that included cyclization of ethyl pyrazolo[1,5-a]pyrimidine-3-carboxylate core, hydrolysis of the ester moiety and amidation (Scheme 1). Specifically, ethyl pyrazolo[1,5-a]pyrimidine-3-carboxylate was synthesized by the reaction of ethyl 3-amino-1H-pyrazole-4-carboxylate with a series of diketone or dialdehyde compounds (1a-n) under acidic conditions. Hydrolysis of these intermediates with sodium hydroxide produced the corresponding carboxylic acids in good yields. For the synthesis of 3a, boration and benzylation were performed prior to saponification of the ethyl ester of 2a. Due to the difficulty of its isolation, (3-(ethoxycarbonyl)pyrazolo[1,5-a]pyrimidin-6-yl)boronic acid (2ab) was used directly for the next step without further purification. To deliver the final target compounds (21-48), the substituted pyrazolo[1,5-a]pyrimidine-3-carboxylic acids were coupled with substituted amines. Compound 47 was obtained by deprotection of Boc in the presence of hydrogen chloride; it was further derivatized into compound 48 through propargylation reactions with propargyl bromide.
Scheme 1.
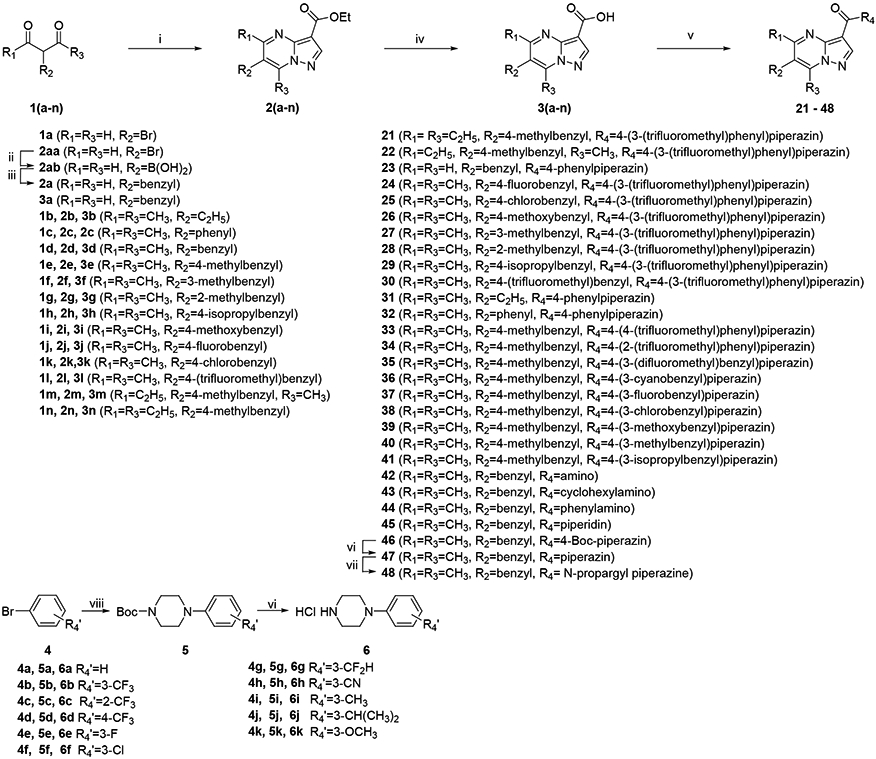
Representative Synthetic Route of pyrazolo[1,5-a]pyrimidine-3-carboxamides. Reagents and conditions: (i) ethyl 3-amino-1H-pyrazole-4-carboxylate, EtOH, AcOH, 70 °C , 3h; (ii) Bis(pinacolato)diboron, KOAc, PdCl2(dppf)(DCM), Dioxane, 80 °C, overnight; (iii) Benzyl bromide, K2CO3, PdCl2(dppf)(DCM), Dioxane, H2O, 50 °C, overnight; (iv) 2 M NaOH, MeOH, THF, 56 °C, 3 h; (v) substituted amines 6a-k, HATU, DIEA, DMF, rt, overnight; (vi) 4M HCl / EtOAc; (vii) propargyl bromide, K2CO3, DCM; (viii) N-Boc piperazine, Pd(OAc)2, XPhos, KOtBu, Dioxane, 90°C, 10h.
2.4. Structure-Activity Relationship of pyrazolo[1,5-a]pyrimidine-3-carboxamide analogs
All of the pyrazolo[1,5-a]pyrimidine-3-carboxamides, including 16-20 from the ChemDiv compound library and the synthesized target compounds 21-48, were evaluated for the inhibitory activity against the RUVBL1/2 complex (Table S1). Among these compounds, Compound 18 showed the most potent inhibition against RUVBL1/2 with an IC50 value of 6.0 ± 0.6 μM. Additionally, 18 could inhibit RUVBL1 with an IC50 value of 7.7 ± 0.9 μM. Methyl groups on R1 and R3 were favorable for inhibitory activity; their replacement by H led to the loss of inhibition (Compound 23). As the volume of the hydrophobic group increases (CH3 → CH2CH3), the inhibitory potency decreases (Compounds 18 → 22 → 21). The substituted benzyl on R2 is essential to the inhibitory activity as the H atom and alkyl groups on R2 resulted in the loss of the inhibitory activity (compounds 20, 31, and 32). The methyl group on the ortho-position of benzyl group led to the loss of the activity against RUVBL1/2 (compound 28). The aromatic-substituted piperazinyl on R4 is essential to the inhibitory activity. However, the heteroaromatic ring-substituted on piperazine decrease the activity compared to the phenyl ring (17 vs 16). When R4 is the amino group substituted by H (compound 42), alky (compounds 43 and 45) or aryl (compound 44) groups, the activity of these compounds decreases greatly. The CF3-substituted on the ortho-position of phenyl ring (on piperazinyl) led to the loss of the activity (compound 34).
2.5. Proposed binding mode of compound 18
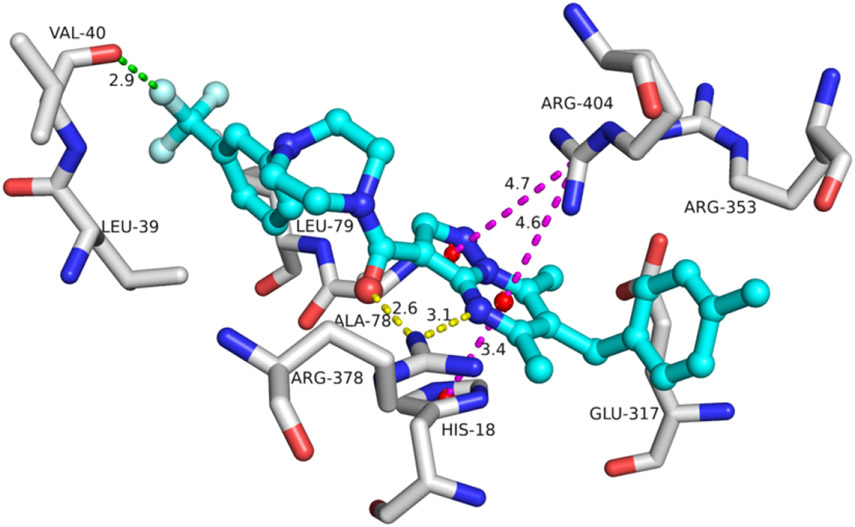
To investigate the binding mode of pyrazolo[1,5-a]pyrimidine-3-carboxamide analogs, we performed a docking study of compound 18 with the RUVBL1/2 complex. Residue Arg378 showed interactions with compound 18 by hydrogen bonds. His18 interacted with the pyrimidine ring of compound 18 by a cation-π interaction. Arg404 showed cation-π interactions with the pyrazolo and pyrimidine of compound 18. There was a potential halogen bond (with 2.9 Å) between the CF3 group and Val40. Multiple residues, such as Leu39, Ala78, Leu79, and the carbon atoms on the side chains of Glu317 and Arg353, showed hydrophobic interactions with these two terminal phenyl rings of compound 18, which is consistent with the study of the structure-activity relationship described above.
2.6. Cytotoxicity of pyrazolo[1,5-a]pyrimidine-3-carboxamide analogs
To determine the cellular activity of pyrazolo[1,5-a]pyrimidine-3-carboxamides, we performed the CellTiter-Glo Luminescent Cell Viability Assay. CB-6644 showed more potent cytotoxicity in the nanomolar IC50 range than did compounds 16-19. Among the pyrazolo[1,5-a]pyrimidine-3-carboxamides, compound 18 was identified as the compound with the most potent cytotoxicity against cell lines A549, HCT116, H1795, and MDA-MB-231 (Table 2 and Figure S1).
Table 2.
The anti-proliferative effects of pyrazolo[1,5-a]pyrimidine-3-carboxamides
| Cmpds | IC50 (μM) |
|||
|---|---|---|---|---|
| A549 | HCT116 | H1795 | MDA-MB-231 | |
| CB-6644 | 0.3 ± 0.07 | 0.08 ± 0.01 | 0.26 ± 0.07 | 0.70 ± 0.29 |
| 16 | 26 ± 5.5 | 21 ± 4.5 | 25 ± 12 | 22 ± 4.3 |
| 17 | 32 ± 1.6 | 32 ± 3.8 | 46 ± 5.5 | 37 ± 8.2 |
| 18 | 15 ± 1.2 | 11 ± 1.0 | 15 ± 1.8 | 8.9 ± 0.9 |
| 19 | 17 ± 1.7 | 17 ± 5.4 | 19 ± 4.1 | 18 ± 3.8 |
2.7. Quantitative Proteomics Analysis of compounds 16, 18, and 19 in treatment of HCT116 human colon cancer cells
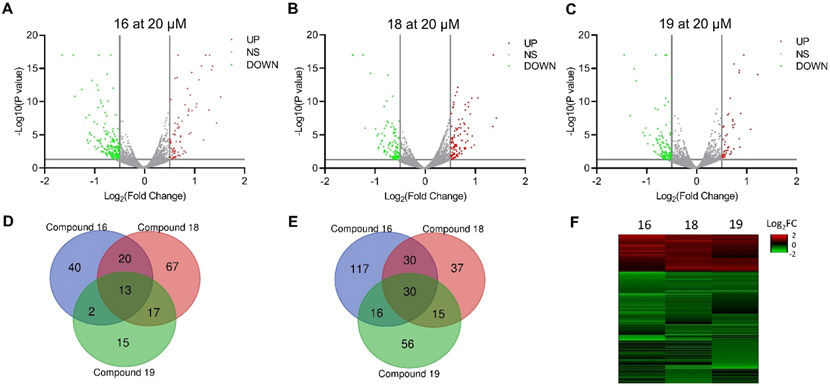
To further study the cellular effects of compounds 16, 18, and 19 and determine which proteins are selectively altered upon treatment, we performed mass spectrometry-based proteomics analysis in HCT116 cells. We evaluated proteome changes using DMSO as negative control after 24 h treatment with either compound 16, 18 or 19. We identified a total of 7693 proteins, among which 268, 229 and 164 were dysregulated by compounds 16, 18 and 19, respectively (P < 0.05, ∣Log2FC∣>0.5) (Figure 5A-C). Of these dysregulated proteins, 193, 112, and 117 were downregulated (Figure 5E). We performed heatmap analysis for differentially expressed proteins identified from two out of three compounds treated groups (p < 0.05, ∣Log2FC∣>0.5) (Figure 5F). The dysregulated proteins from compounds 16, 18, and 19 showed the same trend in the heatmap analysis (Figure 5F). Comparative analysis showed 13 and 30 proteins respectively that were exclusively upregulated and downregulated by pyrazolo[1,5-a]pyrimidine-3-carboxamides 16, 18, and 19 (Figure 5D, Tables S2 and S3).
Figure 5.
Proteomic Effects of Compound Treatment. A-C) Volcano plots of the significantly differentially expressed proteins in HCT116 cells treated with 16, 18 and 19, respectively. D-E) Venn diagrams of upregulated (D) and down-regulated (E) proteins (p < 0.05, ∣Log2FC∣>0.5) following treatment with 16, 18 and 19, respectively. F) Heatmap of differentially regulated proteins from at least two compound-treated groups (p < 0.05, ∣Log2FC∣>0.5).
2.8. GO and KEGG pathway analysis of differentially expressed proteins
To better understand the biological functions of the differentially expressed proteins, we performed GO enrichment and KEGG pathway analyses (Figure 6). The four most significant processes identified by GO analysis annotations of biological process (BP) were regulation of mitotic cell cycle, cell division, cell proliferation, and response to cytokine. The two most significant processes identified among the annotations of cellular component (CC) was nucleus and extracellular space. Two most significant processes identified among annotations of molecular function (MF) were kinase activity and DNA binding. Signaling pathway enrichment analysis of the differentially expressed proteins identified two meaningful pathways with P-value<0.05, namely proteoglycans in cancer and the HIF-1 signaling pathway. Our pathway analysis results were consistent with the various roles of RUVBL1/2 ATPase in regulating cell proliferation, cell cycle [34], regulate chromatin complexes [35] and DNA replication [36]. In our recent review [37], several published studies are referenced to discuss that RUVBL1 and 2 are involved in many cellular activities that are also enriched in our inhibitors induced pathway analysis.
Figure 6.
GO and KEGG pathway enrichment analysis of the proteins dysregulated in at least two compound-treated samples. BP: biological progress; CC: cellular component; MF: molecular function.
3. Conclusion
RUVBL1 and RUVBL2 are well-known drug targets that assemble into heterohexameric rings. Beginning with docking-based virtual screening, 97 compounds were selected and their inhibition activity against RUVBL1/2 complex was evaluated. Seven compounds showed inhibitory activity against the RUVBL1/2 complex in both enzymatic and cellular assays. Due to the chemical space and observed activity, compound 15 was selected as the hit compound for further structural optimization due to its ability to inhibit RUVBL1 and RUVBL2 in addition to the RUVBL1/2 complex. In order to study the structure-activity relationship, a series of pyrazolo[1,5-a]pyrimidine-3-carboxamides (28 synthesized compounds and 10 selected compounds purchased from ChemDiv) were evaluated. Structure-activity relationship analysis suggested that the substituted benzyl on R2 and the aromatic-substituted piperazinyl on R4are essential to the RUVBL1/2 inhibition. Compounds 16, 18, and 19 showed potent inhibition of both enzymatic and cellular activity. The proposed binding mode of compound 18 by docking implied the involvement of hydrophobic interactions between the active residues and the substituents on R2 andR4. MS-based proteomics identified 13 and 30 proteins were up- and down-regulated respectively by pyrazolo[1,5-a]pyrimidine-3-carboxamides 16, 18, and 19. Compound 18 could be used as a lead compound for further therapeutic development.
4. Materials and Methods
4.1. Chemical Synthesis
4.1.1. Materials
All solvents and chemical reagents were purchased from commercial sources and used directly without further purification. The progress of all reactions was monitored by analytical thin-layer chromatography on silica gel 60 F254 plates (Merck). The purification by column chromatography was performed using 200-300 mesh silica gel (Qingdao Haiyang Chemical Co. Ltd.). 1H-NMR spectra were recorded on a Bruker AVANCE 300 or 500 MHz spectrometer in DMSO-d6. 13C-NMR spectra were recorded on a Bruker AVANCE 75 or 125 MHz spectrometer in DMSO-d6. Chemical shift values were reported in δ values parts per million (ppm), referenced to dimethyl sulfoxide (DMSO) using the residual solvent signal as an internal reference. High-resolution mass spectrometry data (HRMS) were recorded on the ICR (Fourier transform ion cyclotron resonance, FTICR) analyzer using an ESI source. Infrared spectra (IR) were recorded on a Thermo Scientific Nicolet iS5 FT-IR spectrophotometer and reported as wavelength numbers (cm−1). Melting points were determined with an X-4A microscopic melting point apparatus, which was uncorrected. All of the test compounds exhibited >95% purity as determined by HPLC. HPLC analysis was performed using an Agilent HPLC 1200 system (column, Agilent C18, 4.6 mm × 150 mm, 5 μm).
4.1.2. General Procedure for the Synthesis of ethyl pyrazolo[1,5-a]pyrimidine-3-carboxylates (2a-n).
The mixture of ethyl 3-amino-1H-pyrazole-4-carboxylate (775 mg, 5.0 mmol, 1.0 eq.) and diketone or dialdehyde compounds (5.25 mmol, 1.05 eq.) in AcOH (4 mL) and EtOH (4 mL) was stirred at 70 °C for 3 h. The mixture was concentrated in vacuo and then partitioned between 50 mL EtOAc and 50 mL NaHCO3 aqueous solution. The organic layer was collected, and the aqueous phase was extracted with 30 mL EtOAc. The combined organic phases were dried over anhydrous Na2SO4 and concentrated in vacuo. The residues were purified by column chromatography (silica gel, petroleum ether (b.p. 60-90 °C)/ethyl acetate, 20:1 to 3:1) to obtain the intermediates.
Ethyl 6-bromopyrazolo[1,5-a]pyrimidine-3-carboxylate (2aa).
Yield: 60%. Yellow solid. M.P. 140.7-142.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 9.75 (d, J = 2.1 Hz, 1 H), 8.89 (d, J = 2.1 Hz, 1 H), 8.59 (s, 1 H), 4.28 (q, J = 7.0 Hz, 2 H), 1.29 (t, J = 7.0 Hz, 3 H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 161.4, 153.7, 147.7, 145.2, 137.4, 105.7, 102.3, 59.8, 14.4. IR νmax (neat): 3062, 2976, 1708, 1539, 1375, 1259, 1209, 1195, 1050, 902, 806, 782, 695. HRMS (ESI+): m/z [2M+Na]+ calcd. for C18H16Br2N6O4Na+, 560.9492; found, 560.9492 (error 0 ppm).
Ethyl 6-benzylpyrazolo[1,5-a]pyrimidine-3-carboxylate (2a).
A mixture of ethyl 6-bromopyrazolo[1,5-a]pyrimidine-3-carboxylate 2aa (2.0 g, 7.4 mmol, 1.0 eq.), Bis(pinacolato)diboron (2.26 g, 8.8 mmol, 1.2 eq.), KOAc (2.18 g, 22.2 mmol, 3.0 eq.) and 1,1'-bis(diphenylphosphino)ferrocene-palladium(II)dichloride complex with DCM (0.42 g, 0.52 mmol, 0.07 eq.) in dioxane (40 mL) was stirred at 80 °C overnight. The reaction mixture was allowed to cool to room temperature and partitioned between 50 mL EtOAc and 50 mL 2M HCl. The aqueous layer was extracted twice with 30 mL EtOAc and the combined organic layers were concentrated in vacuo. The residue was dissolved in 50 mL EtOAc and 50 mL 2M NaOH was added. After separation, the aqueous layer was acidified with hydrochloric acid and extracted with EtOAc (50 mL×3). The combined organic layer was dried over Na2SO4 and concentrated in vacuo to give the crude product 2ab (1.13 g, 4.8 mmol, 65%) as white solid, which was subjected to the next reaction without further purification.
A mixture of (3-(ethoxycarbonyl)pyrazolo[1,5-a]pyrimidin-6-yl)boronic acid 2ab (1.13 g, 4.8 mmol,1.0 eq.), benzyl bromide (0.84 g, 4.8 mmol, 1.0 eq.), and 1,1'-bis(diphenylphosphino)ferrocene-palladium(II)dichloride complex with DCM (0.39 g, 0.48 mmol, 0.1 eq.) in dioxane (30 mL) was treated with a solution of K2CO3 (1.32 g, 9.6 mmol, 2.0 eq.) in H2O (6 mL). The reaction mixture was stirred at 50 °C overnight. The reaction mixture was allowed to cool to room temperature and partitioned between 50 mL EtOAc and 50 mL H2O. The aqueous layer was extracted twice with 30 mL EtOAc. The combined organic layers were dried over anhydrous Na2SO4 followed by filtration and solvent removal in vacuo. The residue was purified by column chromatography (silica gel, petroleum ether (b.p. 60-90 °C)/ethyl acetate, 20:1 to 2:1) to give 2a. (643 mg, 2.28 mmol) Yield: 47%. White solid. M.P. 121.3-123.0 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 9.23 (s, 1H), 8.78 (m, 1H), 8.57 (s, 1H), 7.28-7.35 (m, 4H), 7.19-7.24 (m, 1H), 4.27 (q, J = 7.0 Hz, 2H), 4.08 (s, 2H), 1.28 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 161.6, 155.0, 147.1, 145.7, 139.4, 135.1, 128.7, 128.6, 126.5, 123.9, 101.6, 59.5, 34.5, 14.4. IR νmax (neat): 3051, 2989, 2952, 2930, 1802, 1704, 1629, 1553, 1513, 1389, 1198, 1058, 786, 699. HRMS (ESI+): m/z [2M+Na]+ calcd. for C32H30N6O4Na+, 585.2221; found, 585.2210 (error 1.84 ppm).
Ethyl 6-ethyl-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylate (2b).
Yield: 51%. White solid. M.P. 107.9-109.4 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.46 (s, 1H), 4.26 (q, J = 7.0 Hz, 2H), 2.67-2.73 (m, 5H), 2.60 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H), 1.11 (t, J = 7.4 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 161.8, 161.7, 145.9, 145.2, 143.8, 122.5, 100.6, 59.3, 23.3, 20.7, 14.4, 13.5, 13.0. IR νmax (neat): 2971, 2940, 2879, 1705, 1616, 1546, 1527, 1371, 1191, 1106, 1025, 786, 639. HRMS (ESI+): m/z [2M+Na]+ calcd. for C26H34N6O4Na+, 517.2534; found, 517.2534 (error 0 ppm).
Ethyl 5,7-dimethyl-6-phenylpyrazolo[1,5-a]pyrimidine-3-carboxylate (2c).
Yield: 72%. White solid. M.P. 109.9-110.9 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.58 (s, 1H), 7.49-7.55 (m, 3H), 7.39-7.41 (m, 2H), 4.29 (q, J = 7.0 Hz, 2H), 2.47 (s, 3H), 2.31 (s, 3H), 1.32 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 161.8, 161.0, 146.6, 145.8, 144.3, 134.7, 129.9, 128.9, 128.4, 123.5, 101.0, 59.4, 24.7, 15.0, 14.4. IR νmax (neat): 3112, 3051, 2977, 1693, 1612, 1386, 1373, 1289, 1207, 1108, 779, 705, 649. HRMS (ESI+): m/z [2M+Na]+ calcd. for C34H34N6O4Na+, 613.2534; found, 613.2523 (error 1.75 ppm).
Ethyl 6-benzyl-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylate (2d).
Yield: 92%. White solid. M.P. 170.8-172.3 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.54 (s, 1H), 7.26-7.31 (m, 2H), 7.18-7.22 (m, 1H), 7.11-7.13 (m, 2H), 4.27 (q, J = 7.0 Hz, 2H), 4.19 (s, 2H), 2.78 (s, 3H), 2.48 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.4, 161.8, 146.1, 145.5, 145.2, 138.3, 128.7, 127.9, 126.4, 119.4, 100.9, 59.3, 32.6, 23.8, 14.5, 13.8. IR νmax (neat): 3108, 3083, 3002, 2981, 1679, 1617, 1544, 1196, 1121, 781, 724. HRMS (ESI+): m/z [2M+Na]+ calcd. for C36H38N6O4Na+, 641.2847; found, 641.2847 (error 0 ppm).
Ethyl 5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylate (2e).
Yield: 66%. White solid. M.P. 103.7-105.9 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.51 (s, 1H), 7.06 (d, J = 7.9 Hz, 2H), 6.97 (d, J = 7.9 Hz, 2H), 4.27 (q, J = 7.0 Hz, 2H), 4.10 (s, 2H), 2.75 (s, 3H), 2.46 (s, 3H), 2.22 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.3, 161.8, 146.0, 145.5, 145.0, 135.0, 129.2, 127.6, 119.5, 100.9, 59.2, 32.2, 23.7, 20.5, 14.4, 13.6. IR νmax (neat): 2973, 1720, 1686, 1617, 1550, 1513, 1422, 1370, 1223, 1180, 1107, 974, 838, 780. HRMS (ESI+): m/z [2M+Na]+ calcd. for C38H42N6O4Na+, 669.3160; found, 669.3160 (error 0 ppm).
Ethyl 5,7-dimethyl-6-(3-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylate (2f).
Yield: 82%. White solid. M.P. 115.3-116.7 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.51 (s, 1H), 7.12-7.17 (m, 1H), 6.98-7.01 (m, 1H), 6.87-6.92 (m, 2H), 4.27 (q, J = 7.0 Hz, 2H), 4.12 (s, 2H), 2.76 (s, 3H), 2.47 (s, 3H), 2.22 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.3, 161.8, 146.0, 145.5, 145.1, 138.1, 137.8, 128.5, 128.3, 127.0, 124.9, 119.4, 100.9, 59.3, 32.6, 23.8, 20.9, 14.4, 13.7. IR νmax (neat): 2967, 1712, 1615, 1548, 1372, 1355, 1295, 1246, 1190, 1109, 1027, 975, 836, 792, 690. HRMS (ESI+): m/z [M+H]+ calcd. for C19H22N3O2 +, 324.1707; found, 324.1715 (error −2.61 ppm).
Ethyl 5,7-dimethyl-6-(2-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylate (2g).
Yield: 83%. White solid. M.P. 153.4-154.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.54 (s, 1H), 7.22-7.25 (m, 1H), 7.09-7.14 (m, 1H), 6.97-7.02 (m, 1H), 6.50-6.53 (m, 1H), 4.28 (q, J = 7.0 Hz, 2H), 4.05 (s, 2H), 2.68 (s, 3H), 2.43 (s, 3H), 2.411 (s, 3H), 1.31 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.4, 161.8, 146.0, 145.6, 145.4, 136.1, 130.0, 126.2, 126.1, 125.8, 118.8, 100.9, 59.3, 30.1, 23.4, 19.3, 14.4, 13.5. IR νmax (neat): 2984, 1678, 1618, 1544, 1477, 1381, 1229, 1198, 1122, 1022, 785, 750. HRMS (ESI+): m/z [M+H]+ calcd. for C19H22N3O2+, 324.1707; found, 324.1715 (error −2.61 ppm).
Ethyl 6-(4-isopropylbenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylate (2h).
Yield: 87%. White solid. M.P. 121.5-123.4 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.52 (s, 1H), 7.13 (d, J = 8.0 Hz, 2H), 7.01 (d, J = 8.0 Hz, 2H), 4.27 (q, J = 7.0 Hz, 2H), 4.12 (s, 2H), 2.78-2.87 (m, J = 6.8Hz, 1H), 2.76 (s, 3H), 2.48 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H), 1.14 (d, J = 6.9 Hz, 6H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.3, 161.8, 146.4, 146.1, 145.5, 145.1, 135.5, 127.7, 126.5, 119.5, 100.9, 59.3, 32.9, 32.2, 23.8, 14.4, 13.7. IR νmax (neat): 3107, 2959, 1678, 1614, 1541, 1515, 1379, 1224, 1198, 1124, 1026, 847, 782. HRMS (ESI+): m/z [M+H]+calcd. for C21H26N3O2+, 352.2020; found, 352.2028 (error −2.40 ppm).
Ethyl 6-(4-methoxybenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylate (2i).
Yield: 83%. White solid. M.P. 115.0-116.6 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.53 (s, 1H), 7.03 (d, J = 8.6 Hz, 2H), 6.83 (d, J = 8.6 Hz, 2H), 4.27 (q, J = 7.0 Hz, 2H), 4.10 (s, 2H), 3.70 (s, 3H), 2.77 (s, 3H), 2.48 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.4, 161.8, 157.8, 146.1, 145.5, 145.0, 129.9, 128.9, 119.8, 114.1, 100.8, 59.3, 55.0, 31.8, 23.8, 14.4, 13.7. IR νmax (neat): 2977, 2936, 1720, 1615, 1549, 1514, 1383, 1370, 1116, 1027, 842, 785. HRMS (ESI+): m/z [2M+Na]+ calcd. for C38H42N6O6Na+, 701.3058; found, 701.3058 (error 0 ppm).
Ethyl 6-(4-fluorobenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylate (2j).
Yield: 73%. White solid. M.P. 119.9-122.3 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.52 (s, 1H), 7.14-7.19 (m ,2H), 7.06-7.12 (m, 2H), 4.27 (q, J = 7.0 Hz, 2H), 4.17 (s, 2H), 2.77 (s, 3H), 2.47 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.2, 161.7, 160.7 (d, J = 240.7 Hz), 146.0, 145.4, 145.2, 134.2 (d, J = 2.8 Hz), 129.6 (d, J = 7.9 Hz), 119.2, 115.2 (d, J = 21.0 Hz), 59.2, 31.8, 23.7, 14.4, 13.7. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −116.7. HRMS (ESI+): m/z [M+H]+ calcd. for C18H19FN3O2+, 328.1456; found, 328.1463 (error −2.19 ppm).
Ethyl 6-(4-chlorobenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylate (2k).
Yield: 83%. White solid. M.P. 137.8-140.0 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.52 (s, 1H), 7.32 (d, J = 8.3 Hz, 2H), 7.15 (d, J = 8.3 Hz, 2H), 4.27 (q, J = 7.0 Hz, 2H), 4.17 (s, 2H), 2.76 (s, 3H), 2.46 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.2, 161.8, 146.1, 145.6, 145.4, 137.3, 131.0, 129.7, 128.5, 119.0, 100.9, 59.3, 32.0, 23.8, 14.4, 13.7. IR νmax (neat): 2983, 2897, 1709, 1614, 1550, 1372, 1292, 1242, 1187, 1109, 984, 796, 783. HRMS (ESI+): m/z [2M+Na]+ calcd. for C36H36Cl2N6O4Na+, 709.2067; found, 709.2067 (error 0 ppm).
Ethyl 5,7-dimethyl-6-(4-(trifluoromethyl)benzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylate (2l).
Yield: 84%. White solid. M.P. 143.8-144.4 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.54 (s, 1H), 7.63 (d, J = 8.0 Hz, 2H), 7.37 (d, J = 8.0 Hz, 2H), 4.24-4.30 (m, 4H), 2.78 (s, 3H), 2.47 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.2, 161.7, 146.1, 145.6, 145.5, 143.3, 128.7, 127.1 (q, J = 31.5 Hz), 125.3 (q, J = 3.5 Hz), 124.2 (q, J = 270.2 Hz), 118.5, 100.9, 59.2, 32.5, 23.8, 14.4, 13.7. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −60.9. IR νmax (neat): 2984, 1714, 1616, 1551, 1522, 1374, 1320, 1244, 1163, 1108, 813, 783. HRMS (ESI+): m/z [M+H]+ calcd. for C19H19F3N3O2+, 378.1424; found, 378.1433 (error −2.41 ppm).
Ethyl 5-ethyl-7-methyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylate (2m).
Yield: 58%. White solid. M.P. 121.3-124.1 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.54 (s, 1H), 7.09 (d, J = 7.8 Hz, 2H), 6.98 (d, J = 7.8 Hz, 2H), 4.27 (q, J = 7.0 Hz, 2H), 4.14 (s, 2H), 3.20 (q, J = 7.3 Hz, 2H), 2.46 (s, 3H), 2.24 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H), 1.14 (t, J = 7.3 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm):162.9, 161.7, 149.3, 146.1, 145.6, 135.3, 129.2, 127.6, 118.6, 100.8, 59.2, 31.8, 23.8, 20.6, 20.4, 14.4, 10.9. IR νmax (neat): 2969, 2936, 2873, 1716, 1609, 1548, 1514, 1370, 1347, 1291, 1190, 1121, 1090, 788. HRMS (ESI+): m/z [M+H]+ calcd. for C20H24N3O2+, 338.1863; found, 338.1870 (error −2.06 ppm).
Ethyl 5,7-diethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylate (2n).
Yield: 55%. White solid. M.P. 82.9-84.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.54 (s, 1H), 7.07 (d, J = 7.8 Hz, 2H), 6.96 (d, J = 7.8 Hz, 2H), 4.26 (q, J = 7.0 Hz, 2H), 4.15 (s, 2H), 3.19 (q, J = 7.2 Hz, 2H), 2.76 (q, J = 7.2 Hz, 2H), 2.23 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H), 1.11-1.20 (m, 6H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 166.1, 161.8, 149.2, 146.1, 145.8, 135.6, 135.3, 129.1, 127.5, 118.1, 101.0, 59.2, 31.2, 28.3, 20.7, 20.5, 14.3, 11.3, 10.9. IR νmax (neat): 2969, 1716, 1611, 1544, 1513, 1467, 1369, 1349, 1237, 1120, 1091, 928, 911, 795, 780. HRMS (ESI+): m/z [M+H]+ calcd. for C21H26N3O2+, 352.2020; found, 352.2019 (error 0.15 ppm).
4.1.3. General Procedure for the Synthesis of pyrazolo[1,5-a]pyrimidine-3-carboxylic acids (3a-n)
To a solution of ethyl pyrazolo[1,5-a]pyrimidine-3-carboxylates (4.0 mmol, 1.0 eq.) in THF (3.0 mL) and MeOH (3.0 mL) was added 2 M NaOH aqueous solution (3.0 mL, 6.0 mmol, 1.5 eq.). The mixture was stirred at 56 °C for 3 h and then concentrated in vacuo. The residue was suspended in water, and 2 M HCl aqueous solution (3.0 mL, 6.0 mmol, 1.5 eq.) was added. The resulting solid was collected by filtration, rinsed with water, and dried to afford the intermediates as off-white to yellowish solids.
6-benzylpyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3a).
Yield: 81%. Pale yellow solid. M.P. 237.3-240.1 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 12.39 (s, 1H), 9.20 (s, 1H), 8.74 (s, 1H), 8.54 (s, 1H), 7.28-7.33 (m, 4H), 7.21-7.23 (m, 1H), 4.08 (s, 2H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.1, 154.7, 147.4, 145.8, 139.5, 135.0, 128.7, 128.6, 126.6, 123.7, 102.4, 34.5. IR νmax (neat): 3079, 2921, 1698, 1626, 1558, 1521, 1413, 1386, 1269, 1196, 1049, 778, 721, 641. HRMS (ESI−): m/z [2M-H]− calcd. for C28H21N6O4−, 505.1630; found, 505.1620 (error 1.94 ppm).
6-ethyl-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3b).
Yield: 88%. Pale yellow solid. M.P. 179.5-180.9 °C. 1H NMR (500 MHz, DMSO-d6) δ (ppm): 12.11 (s, 1H), 8.43 (s, 1H), 2.70-2.74 (m, 5H), 2.61 (s, 3H), 1.13 (t, J = 7.5 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 163.2, 161.2, 146.2, 145.1, 143.7, 122.2, 101.3, 23.1, 20.6, 13.4, 12.9. IR νmax (neat): 2972, 2579, 1717, 1643, 1613, 1538, 1523, 1361, 1232, 1190, 784, 733, 657, 640. HRMS (ESI+): m/z [2M+Na]+ calcd. for C22H26N6O4Na+, 461.1908; found, 461.1908 (error 0 ppm).
5,7-dimethyl-6-phenylpyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3c).
Yield: 82%. White solid. M.P. 220.0-221.9 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 12.31 (brs, 1H), 8.54 (s, 1H), 7.45-7.56 (m, 3H), 7.38-7.41 (m, 2H), 2.47 (s, 3H), 2.30 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.4, 160.6, 147.0, 145.8, 144.3, 134.9, 129.9, 128.9, 128.4, 123.2, 101.9, 24.6, 15.0. IR νmax (neat): 3061, 2697, 2631, 2577, 1714, 1658, 1610, 1513, 1386, 1233, 1198, 788, 702. HRMS (ESI+): m/z [2M+Na]+ calcd. for C30H26N6O4Na+, 557.1908; found, 557.1908 (error 0 ppm).
6-benzyl-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3d).
Yield: 85%. White solid. M.P. 180.5-182.9 °C. 1H NMR (500 MHz, DMSO-d6) δ (ppm): 12.16 (s, 1H), 8.49 (s, 1H), 7.26-7.29 (m, 2H), 7.19 (t, J = 7.3 Hz, 1H), 7.11-7.12 (m, 2H), 4.18 (s, 2H), 2.77 (s, 3H), 2.47 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 163.3, 162.0, 146.4, 145.5, 145.1, 138.3, 128.6, 127.8, 126.3, 119.2, 101.6, 32.6, 23.7, 13.7. IR νmax (neat): 2945, 1727, 1660, 1615, 1553, 1493, 1362, 1248, 1198, 784, 736, 701. HRMS (ESI−): m/z [M-H]− calcd. for C16H14N3O2−, 280.1092; found, 280.1092 (error 0 ppm).
5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3e).
Yield: 90%. White solid. M.P. 192.0-193.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 12.58 (brs, 1H), 8.48 (s, 1H), 7.07 (d, J = 7.7 Hz, 2H), 6.98 (d, J = 7.7 Hz, 2H), 4.11 (s, 2H), 2.76 (s, 3H), 2.46 (s, 3H), 2.23 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.8, 161.8, 146.4, 145.3, 144.9, 135.3, 135.1, 129.2, 127.6, 119.1, 102.4, 32.2, 23.6, 20.5, 13.7. IR νmax (neat): 2921, 1660, 1616, 1548, 1515, 1493, 1386, 1360, 1248, 1195, 944, 785. HRMS (ESI−): m/z [M-H]− calcd. for C17H16N3O2 −, 294.1248; found, 294.1253 (error −1.70 ppm).
5,7-dimethyl-6-(3-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3f).
Yield: 84%. White solid. M.P. 180.5-183.1 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 12.37 (brs, 1H), 8.49 (s, 1H), 7.16 (m, 1H), 6.99-7.01 (m, 1H), 6.88-6.93 (m, 2H), 4.13 (s, 2H), 2.77 (s, 3H), 2.47 (s, 3H), 2.23 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.5, 162.0, 146.4, 145.4, 145.0, 138.2, 137.8, 128.5, 128.3, 127.0, 124.9, 119.1, 101.9, 32.6, 23.7, 20.9, 13.7. IR νmax (neat): 3391, 2941, 1672, 1615, 1548, 1521, 1486, 1374, 1231, 1193, 1143, 982, 785, 732. HRMS (ESI+): m/z [M+H]+ calcd. for C17H18N3O2+, 296.1394; found, 296.1394 (error 0 ppm).
5,7-dimethyl-6-(2-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3g).
Yield: 96%. White solid. M.P. >300 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 12.60 (brs, 1H), 8.49 (s, 1H), 7.22-7.25 (m, 1H), 7.11 (m, 1H), 6.99 (m, 1H), 6.49-6.52 (m, 1H), 4.05 (s, 2H), 2.68 (s, 3H), 2.43 (s, 3H), 2.40 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.8, 161.9, 146.4, 145.5, 145.2, 136.2, 136.1, 130.0, 126.2.126.1, 125.8, 118.3, 102.6, 30.0, 23.2, 19.3, 13.5. IR νmax (neat): 3479, 3020, 1654, 1618, 1551, 1532, 1373, 1196, 1137, 981, 845, 789, 741. HRMS (ESI−): m/z [M-H]− calcd. for C17H16N3O2−, 294.1248; found, 294.1237 (error 3.74 ppm).
6-(4-isopropylbenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3h).
Yield: 95%. White solid. M.P. 210.3-211.9 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 12.61 (brs, 1H), 8.48 (s, 1H), 7.13 (d, J = 7.9 Hz, 2H), 7.01 (d, J = 7.9 Hz, 2H), 4.12 (s, 2H), 2.76-2.85 (m, 4H), 2.47 (s, 3H), 1.13 (d, J = 6.8 Hz, 6H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.8, 161.8, 146.4, 146.3, 145.3, 144.9, 135.5, 127.7, 126.5, 119.1, 102.6, 32.9, 32.2, 23.8, 23.6, 13.7. IR νmax (neat): 3464, 2957, 1683, 1619, 1548, 1523, 1377, 1364, 1299, 1192, 1144, 849, 787, 736. HRMS (ESI+): m/z [2M+Na]+ calcd. for C38H42N6O4Na+, 669.3160; found, 669.3168 (error −1.23 ppm).
6-(4-methoxybenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3i).
Yield: 91%. White solid. M.P. 213.2-214.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 12.25 (brs, 1H), 8.49 (s, 1H), 7.03 (d, J = 8.5 Hz, 2H), 6.83 (d, J = 8.5 Hz, 2H), 4.10 (s, 2H), 3.70 (s, 3H), 2.77 (s, 3H), 2.47 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.3, 162.0, 157.8, 146.4, 145.4, 144.9, 130.0, 128.8, 119.5, 114.0, 101.7, 55.0, 31.8, 23.6, 13.7. IR νmax (neat): 2937, 1661, 1611, 1548, 1511, 1426, 1246, 1190, 1021, 844, 785, 744. HRMS (ESI+): m/z [M+H]+ calcd. for C17H18N3O3+, 312.1343; found, 312.1348 (error −1.70 ppm).
6-(4-fluorobenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3j).
Yield: 96%. White solid. M.P. 201.0-202.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.48 (s, 1H), 7.06-7.18 (m, 4H), 4.16 (s, 2H), 2.77 (s, 3H), 2.46 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.8, 161.6, 160.7 (d, J = 240.8 Hz), 146.4, 145.3, 145.0, 134.4 (d, J = 2.8 Hz), 129.6 (d, J = 7.8 Hz), 118.8, 115.2 (d, J = 21.0 Hz), 102.7, 31.8, 23.6, 13.7. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −116.7. IR νmax (neat): 3063, 2585, 1659, 1615, 1550, 1509, 1385, 1362, 1249, 1196, 945, 852, 786, 770, 747. HRMS (ESI+): m/z [2M+Na]+ calcd. for C32H28F2N6O4Na+, 621.2032; found, 621.2032 (error 0 ppm).
6-(4-chlorobenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3k).
Yield: 95%. White solid. M.P. >300 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 12.52 (brs, 1H), 8.49 (s, 1H), 7.33 (d, J = 8.3 Hz, 2H), 7.16 (d, J = 8.3 Hz, 2H), 4.18 (s, 2H), 2.77 (s, 3H), 2.46 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.7, 161.6, 146.5, 145.4, 145.2, 137.4, 130.9, 129.7, 128.5, 118.5, 102.7, 32.0, 23.6, 13.7. IR νmax (neat): 3228, 1615, 1549, 1525, 1386, 1373, 1247, 1189, 1093, 1013, 908, 848, 797, 744. HRMS (ESI−): m/z [M-H]− calcd. for C16H13ClN3O2 −, 314.0702; found, 314.0691 (error 3.44 ppm).
5,7-dimethyl-6-(4-(trifluoromethyl)benzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3l).
Yield: 90%. White solid. M.P. >300 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.46 (s, 1H), 7.63 (d, J = 7.8 Hz, 2H), 7.36 (d, J = 7.8 Hz, 2H), 4.30 (s, 2H), 2.77 (s, 3H), 2.47 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 161.2, 146.5, 145.2, 143.5, 128.6, 127.0 (q, J = 31.3 Hz), 125.4 (q, J = 3.6 Hz), 124.2 (q, J = 270.1 Hz), 117.8, 32.5, 23.6, 13.7. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −60.8. IR νmax (neat): 3475, 1615, 1551, 1407, 1375, 1328, 1186, 1109, 1067, 902, 808. HRMS (ESI−): m/z [M-H]− calcd. for C17H13F3N3O2−, 348.0965; found, 348.0964 (error 0.39 ppm).
5-ethyl-7-methyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3m).
Yield: 91%. White solid. M.P. >300 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.27 (s, 1H), 7.07 (d, J = 7.5 Hz, 2H), 6.95 (d, J = 7.5 Hz, 2H), 4.10 (s, 2H), 3.19 (q, J = 7.2 Hz, 2H), 2.43 (s, 3H), 2.24 (s, 3H), 1.14 (t, J = 7.2 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 148.4, 146.4, 144.5, 135.8, 135.2, 129.1, 127.5, 116.5, 31.8, 23.4, 20.5, 11.0. IR νmax (neat): 3374, 2936, 1613, 1544, 1514, 1408, 1389, 1308, 1189, 808, 772. HRMS (ESI+): m/z [2M+Na]+ calcd. for C36H38N6O4Na+, 641.2847; found, 641.2847 (error 0 ppm).
5,7-diethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carboxylic acid (3n).
Yield: 80%. Faint yellow solid. M.P. 158.1-160.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 12.13 (brs, 1H), 8.51 (s, 1H), 7.08 (d, J = 7.7 Hz, 2H), 6.97 (d, J = 7.7 Hz, 2H), 4.15 (s, 2H), 3.19 (q, J = 7.1 Hz, 2H), 2.75 (q, J = 7.3 Hz, 2H), 2.24 (s, 3H), 1.12-1.17 (m, 6H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 166.1, 163.2, 149.3, 146.6, 145.8, 135.7, 135.3, 129.2, 127.6, 117.8, 101.7, 31.2, 28.4, 20.7, 20.5, 11.8, 10.9. IR νmax (neat): 3451, 2983, 2939, 1666, 1617, 1543, 1372, 1236, 1193, 1148, 787, 736. HRMS (ESI+): m/z [M+H]+ calcd. for C19H22N3O2+, 324.1707; found, 324.1714 (error −2.30 ppm).
4.1.4. General Procedure for the Synthesis of tert-butyl 4-phenylpiperazine-1-carboxylates (5a-k).
A mixture of N-Boc piperazine (5.0 mmol, 1.0 eq.) and substituted bromo benzenes 4 (5.5 mmol, 1.1 eq.) in the presence of palladium acetate (0.5 mmol, 0.1 eq.), 2-Dicyclohexylphosphino-2',4',6'-triisopropylbiphenyl (1.0 mmol, 0.2 eq.) and potassium t-butoxide (6.0 mmol, 1.2 eq.) in dioxane (15 ml) was stirred in at 90 °C for 10 h. The reaction mixture was allowed to cool to room temperature, and 100 ml ethyl acetate and 100 ml water were added. The thus-obtained mixture was filtered through a celite pad. The organic layer was collected, and the aqueous phase was extracted with 50 mL EtOAc. The combined organic phases were dried over anhydrous Na2SO4 and concentrated in vacuo. The residues were purified by column chromatography (silica gel, petroleum ether (b.p. 60-90 °C)/ethyl acetate, 100:1 to 20:1) to afford the intermediates.
tert-Butyl 4-phenylpiperazine-1-carboxylate (5a).
Yield: 40%. Light brown solid. M.P. 68.0-69.1 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.19-7.24 (m, 2H), 6.92-6.95 (m, 2H), 6.80 (t, J = 7.2 Hz, 1H), 3.43-3.45 (m, 4H), 3.05-3.08 (m, 4H), 1.42 (s, 9H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 153.8, 150.9, 128.9, 119.3, 116.0, 78.9, 48.4, 42.8 (br), 28.0. IR νmax (neat): 2979, 2929, 2859, 1684, 1597, 1427, 1389, 1364, 1235, 1156, 753, 689. HRMS (ESI+): m/z [M+H]+ calcd. for C15H23N2O2+, 263.1754; found, 263.1754 (error 0 ppm).
tert-Butyl 4-(3-(trifluoromethyl)phenyl)piperazine-1-carboxylate (5b).
Yield: 37%. Light brown solid. M.P. 45.3-47.6 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.39-7.41 (m, J = 7.9 Hz, 1H), 7.17-7.22 (m, 2H), 7.06-7.09 (m, 1H), 3.44-3.46 (m, 4H), 3.18-3.20 (m, 4H), 1.42 (s, 9H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 153.8, 151.1, 129.9 (q, J = 30.8 Hz), 129.8, 124.3 (q, J = 270.7 Hz), 119.1, 114.9 (q, J = 3.7 Hz), 111.3 (q, J = 3.7 Hz), 78.9, 47.6, 43.0 (br), 28.0. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −61.1. IR νmax (neat): 2979, 2933, 2828, 1684, 1607, 1420, 1389, 1362, 1234, 1153, 1109, 942, 857, 782, 691. HRMS (ESI+): m/z [M+H]+ calcd. for C16H22F3N2O2+, 331.1628; found, 331.1628 (error 0 ppm).
tert-Butyl 4-(2-(trifluoromethyl)phenyl)piperazine-1-carboxylate (5c).
Yield: 51%. White solid. M.P. 46.8-48.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.62-7.67 (m, 2H), 7.54-7.56 (m, 1H), 7.32-7.37 (m, 1H), 3.43 (br, 4H), 2.78-2.81 (m, 4H), 1.42 (s, 9H). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 153.8, 152.0, 133.6, 126.8 (q, J = 5.3 Hz), 125.7 (q, J = 27.7 Hz), 125.5, 124.7, 124.0 (q, J = 271.5 Hz), 78.9, 52.9, 44.1 (br), 43.3 (br), 28.0. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −58.8. IR νmax (neat): 2979, 2858, 1682, 1424, 1388, 1366, 1311, 1248, 1123, 780, 767. HRMS (ESI+): m/z [M+Na]+ calcd. for C16H21F3N2O2Na+, 353.1447; found, 353.1442 (error 1.51 ppm).
tert-Butyl 4-(4-(trifluoromethyl)phenyl)piperazine-1-carboxylate (5d).
Yield: 63%. Light brown solid. M.P. 125.9-127.0 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.50 (d, J = 8.7 Hz, 2H), 7.05 (d, J = 8.7 Hz, 2H), 3.43-3.47 (m, 4H), 3.23-3.27 (m, 4H), 1.41 (s, 9H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 153.9, 153.1, 126.1 (q, J = 3.5 Hz), 124.9 (q, J = 268.6 Hz), 118.1 (q, J = 31.8 Hz), 114.5, 79.1, 46.9, 42.8 (br), 28.0. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −59.5. IR νmax (neat): 3001, 2931, 2820, 1672, 1614, 1418, 1392, 1367, 1326, 1232, 1137, 1107, 831, 632. HRMS (ESI+): m/z [M+H]+ calcd. for C16H22F3N2O2+, 331.1628; found, 331.1623 (error 1.47 ppm).
tert-Butyl 4-(3-fluorophenyl)piperazine-1-carboxylate (5e).
Yield: 48%. White solid. M.P. 44.0-45.7 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.17-7.25 (m, 1H), 6.71-6.76 (m, 2H), 6.53-6.58 (m, 1H), 3.42-3.43 (m, 4H), 3.11-3.14 (m, 4H), 1.41 (s, 9H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.1 (d, J = 239.0 Hz), 153.8, 152.5 (d, J = 10.0 Hz), 130.3 (d, J = 10.0 Hz), 111.2, 104.9 (d, J = 21.2 Hz), 102.1 (d, J = 25.0 Hz), 78.9, 47.6, 42.9 (br), 28.0. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −112.3. IR νmax (neat): 2981, 2859, 1682, 1609, 1581, 1497, 1424, 1390, 1364, 1243, 1157, 1124, 996, 967, 759, 682. HRMS (ESI+): m/z [M+H]+ calcd. for C15H22FN2O2+, 281.1660; found, 281.1660 (error 0 ppm).
tert-Butyl 4-(3-chlorophenyl)piperazine-1-carboxylate (5f).
Yield: 51%. White solid. M.P. 64.6-65.8 °C. 1H NMR (500 MHz, DMSO-d6) δ (ppm): 7.19-7.22 (m, 1H), 6.94-6.95 (m, 1H), 6.87-6.89 (m, 1H), 6.78-6.80 (m, 1H), 3.42-3.44 (m, 4H), 3.11-3.13 (m, 4H), 1.41 (s, 9H). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 153.8, 152.1, 133.8, 130.4, 118.5, 115.0, 114.1, 79.0, 47.7, 43.2 (br), 28.0. IR νmax (neat): 2976, 2861, 1679, 1592, 1424, 1391, 1364, 1235, 1158, 1124, 933, 760, 679. HRMS (ESI+): m/z [M+H]+ calcd. for C15H22ClN2O2+, 297.1364; found, 297.1365 (error −0.23 ppm).
tert-Butyl 4-(3-(difluoromethyl)phenyl)piperazine-1-carboxylate (5g).
Yield: 51%. Light brown solid. M.P. 61.1-62.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.32-7.37 (m, 1H), 7.10 (m, 2H), 6.97-6.99 (m, 1H), 6.91 (t, J = 56.0 Hz, 1H), 3.44-3.46 (m, 4H), 3.13-3.16 (m, 4H), 1.42 (s, 9H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 153.8, 151.0, 134.9 (t, J = 21.6 Hz), 129.5, 117.9, 115.9 (t, J =5.8 Hz), 115.2 (t, J = 234.6 Hz), 112.6 (t, J = 6.3 Hz), 79.0, 47.9, 43.0 (br), 28.0. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −112.3. IR νmax (neat): 2977, 2862, 1682, 1425, 1388, 1364, 1244, 1165, 1129, 1018, 950, 867, 801, 751. HRMS (ESI+): m/z [M+H]+ calcd. for C16H23F2N2O2+, 313.1722; found, 313.1720 (error 0.67 ppm).
tert-Butyl 4-(3-cyanophenyl)piperazine-1-carboxylate (5h).
Yield: 28%. White solid. M.P. 130.1-131.2 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.33-7.41 (m, 2H), 7.25-7.28 (m, 1H), 7.15-7.18 (m, 1H), 3.42-3.44 (m, 4H), 3.17-3.20 (m, 4H), 1.41 (s, 9H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 153.8, 150.9, 130.1, 121.9, 120.0, 119.2, 118.0, 112.0, 79.0, 47.3, 42.8 (br), 28.0. IR νmax (neat): 2979, 2866, 2222, 1674, 1598, 1573, 1425, 1393, 1364, 1241, 1157, 1123, 992, 952, 783, 683. HRMS (ESI+): m/z [M+H]+ calcd. for C16H22N3O2+, 288.1707; found, 288.1709 (error −0.86 ppm).
tert-Butyl 4-(m-tolyl)piperazine-1-carboxylate (5i).
Yield: 51%. Light brown solid. M.P. 66.3-67.1 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.07-7.12 (m, 1H), 6.71-6.75 (m, 2H), 6.61-6.63 (m, 1H), 3.42-3.45 (m, 4H), 3.03-3.06 (m, 4H), 2.25 (s, 3H), 1.42 (s, 9H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 153.8, 150.9, 137.9, 128.7, 120.1, 116.6, 113.1, 78.8, 48.4, 43.2 (br), 28.0, 21.3. IR νmax (neat): 2973, 2917, 2820, 1694, 1599, 1423, 1385, 1363, 1421, 1157, 1118, 948, 767, 688. HRMS (ESI+): m/z [M+H]+ calcd. for C16H25N2O2+, 277.1911; found, 277.1911 (error 0 ppm).
tert-Butyl 4-(3-isopropylphenyl)piperazine-1-carboxylate (5j).
Yield: 72%. White solid. M.P. 39.7-40.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.10-7.15 (m, 1H), 6.81 (s, 1H), 6.68-6.75 (m, 2H), 3.43-3.45 (m, 4H), 3.04-3.07 (m, 4H), 2.81 (hept, J = 6.8 Hz, 1H), 1.42 (s, 9H), 1.18 (d, J = 6.8 Hz, 6H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 153.8, 151.0, 149.1, 128.7, 117.4, 114.3, 113.5, 78.8, 48.6, 43.1 (br), 33.7, 28.0, 23.9. IR νmax (neat): 2958, 2861, 2822, 1687, 1605, 1421, 1364, 1339, 1213, 1160, 1121, 953, 778, 703. HRMS (ESI+): m/z [M+H]+ calcd. for C18H29N2O2+, 305.2224; found, 305.2224 (error 0 ppm).
tert-Butyl 4-(3-methoxyphenyl)piperazine-1-carboxylate (5k).
Yield: 36%. White solid. M.P. 59.8-61.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 7.09-7.14 (m, 1H), 6.38-6.53 (m, 3H), 3.71 (s, 3H), 3.42-3.45 (m,4H), 3.05-3.09 (m, 4H), 1.42 (s, 9H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 160.2, 153.8, 152.2, 129.6, 108.5, 104.6, 102.0, 78.9, 54.8, 48.3, 43.1 (br), 28.0. IR νmax (neat): 2973, 2923, 2873, 1690, 1585, 1417, 1369, 1245, 1162, 995, 945, 819, 760, 684. HRMS (ESI+): m/z [M+H]+ calcd. for C16H25N2O3+, 293.1860; found, 293.1858 (error 0.58 ppm).
4.1.5. General Procedure for the Synthesis of Pyrazolo[1,5-a]pyrimidine-3-carboxamides (21-48).
The Boc protected 4-phenylpiperazines (1.5 mmol) were dissolved in 8 ml of 4M HCl/EtOAc and the reaction mixture was stirred at room temperature for 2 h. Then the reaction mixture was evaporated in vacuo to give the crude piperazine hydrochlorides as near white solid, which was subjected to the next reaction without further purification.
To a mixture of pyrazolo[1,5-a]pyrimidine-3-carboxylic acids (1.0 mmol, 1.0 eq.) and amine (1.1 mmol, 1.1 eq.) in DMF (8 mL) was added HATU (1.5 mmol, 1.5 eq.), followed by DIEA (3.0 mmol, 3.0 eq.). The mixture was stirred at room temperature overnight and then diluted with 80 mL EtOAc and washed with 30 mL saturated aqueous NaCl. The organic layer was dried over anhydrous Na2SO4 and concentrated in vacuo. The residue was purified by column chromatography (silica gel, petroleum ether (b.p.60-90°C)/ethyl acetate, 20:1 to 1:3) to afford the desired compounds as solid.
(5,7-diethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (21).
Yield: 87%. White solid. M.P. 64.4-66.0 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.40 (s, 1H), 7.40-7.45 (m, 1H), 7.21-7.25 (m, 2H), 7.08-7.10 (m, 3H), 6.98-7.01 (m, 2H), 4.15 (s, 2H), 3.78 (s, 4H), 3.34 (s, 4H), 3.21 (q, J = 7.4 Hz, 2H), 2.75 (q, J = 7.3 Hz, 2H), 2.24 (s, 3H), 1.13-1.20 (m, 6H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 164.7, 162.7, 151.1, 148.9, 145.8, 142.7, 135.7, 135.3, 130.0, 129.9 (q, J = 30.8 Hz), 129.2, 127.6, 124.3 (q, J = 270.8 Hz), 119.0, 117.0, 114.9 (q, J = 3.6 Hz), 111.2 (q, J = 3.7 Hz), 104.8, 48.2 (br), 31.2, 28.3, 20.7, 20.5, 11.5, 10.9. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −61.1. IR νmax (neat): 2935, 1732, 1610, 1544, 1514, 1449, 1350, 1306, 1232, 1158, 1116, 994, 944, 844, 786, 695. HRMS (ESI+): m/z [M+H]+ calcd. for C30H33F3N5O+, 536.2632; found, 536.2632 (error 0 ppm).
(5-ethyl-7-methyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (22).
Yield: 84%. White solid. M.P. 62.3-63.7 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.39 (s, 1H), 7.39-7.44 (m, 1H), 7.21-7.24 (m, 2H), 7.07-7.10 (m, 3H), 6.98-7.01 (m, 2H), 4.12 (s, 2H), 3.75 (s, 4H), 3.34 (m, 4H), 3.21 (q, J = 7.2 Hz, 2H), 2.43 (s, 3H), 2.23 (s, 3H), 1.18 (t, J = 7.2 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.6, 161.1, 151.0, 148.8, 145.4, 142.8, 135.5, 135.3, 130.1, 129.9 (q, J = 30.8 Hz), 129.2, 127.6, 124.3 (q, J = 270.7 Hz), 118.9, 117.4, 114.8 (q, J = 3.6 Hz), 111.2 (q, J = 3.7 Hz), 104.6, 48.0 (br), 31.8, 23.7, 20.6, 20.4, 10.9. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −61.1. IR νmax (neat): 2938, 1613, 1547, 1514, 1449, 1391, 1318, 1306, 1232, 1159, 1116, 993, 944, 857, 782, 696. HRMS (ESI+): m/z [M+Na]+ calcd. for C29H30F3N5ONa+, 544.2295; found, 544.2292 (error 0.49 ppm).
(6-benzylpyrazolo[1,5-a]pyrimidin-3-yl)(4-phenylpiperazin-1-yl)methanone (23).
Yield: 85%. White solid. M.P. 120.7-122.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 9.18 (s, 1H), 8.66 (m, 1H), 8.41 (s, 1H), 7.29-7.38 (m, 4H), 7.18-7.23 (m, 3H), 6.92-6.95 (m, 2H), 6.77-6.81 (m, 1H), 4.06 (s, 2H), 3.70 (brs, 4H), 3.17 (brs, 4H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.1, 153.5, 150.8, 146.0, 143.0, 139.6, 134.5, 128.9, 128.7, 126.5, 122.9, 119.3, 115.9, 105.4, 48.8, 34.5. IR νmax (neat): 3059, 2919, 2853, 1669, 1627, 1494, 1384, 1231, 1205, 1007, 842, 761, 698. HRMS (ESI+): m/z [2M+H]+ calcd. for C48H47N10O2+, 795.3878; found, 795.3876 (error 0.25 ppm).
(6-(4-fluorobenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (24).
Yield: 69%. White solid. M.P. 60.6-62.2 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.39 (s, 1H), 7.40-7.45 (m, 1H), 7.17-7.25 (m, 4H), 7.07-7.13 (m, 3H), 4.17 (s, 2H), 3.75 (s, 4H), 3.34 (s, 4H), 2.79 (s, 3H), 2.45 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.6, 160.8 (d, J = 240.8 Hz), 160.6, 151.0, 145.4, 144.7, 142.7, 134.5 (d, J = 2.8 Hz), 130.0, 129.9 (q, J = 30.7 Hz), 129.7, 129.6, 124.3 (q, J = 270.8 Hz), 118.9, 118.1, 115.4, 115.1, 114.8 (q, J = 3.7 Hz), 111. 2 (q, J = 3.7 Hz), 104.6, 48.0 (br), 31.8, 23.7, 13.6. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −61.1, −116.8. IR νmax (neat): 2916, 1731, 1614, 1548, 1507, 1429, 1352, 1318, 1306, 1220, 1156, 1115, 989, 943, 845, 784, 695. HRMS (ESI+): m/z [M+H]+ calcd. for C27H26F4N5O+, 512.2068; found, 512.2062 (error 1.17 ppm).
(6-(4-chlorobenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (25).
Yield: 67%. White solid. M.P. 180.1-181.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.39 (s 1H), 7.40-7.45 (m, 1H), 7.32-7.35 (m, 2H), 7.17-7.26 (m, 4H), 7.07-7.09 (m, 1H), 4.18 (s, 2H), 3.74 (s, 4H), 3.35 (s, 4H), 2.78 (s, 3H), 2.44 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.6, 160.6, 151.1, 145.4, 144.9, 142.8, 137.6, 130.9, 130.0, 129.9 (q, J = 31.0 Hz), 129.7, 128.5, 124.4 (q, J = 270.7 Hz), 119.0, 117.9, 114.8 (q, J = 3.6 Hz), 111.3 (q, J = 3.6 Hz), 104.7, 48.1 (br), 32.0, 23.8, 13.7. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −61.1. IR νmax (neat): 3103, 2816, 1608, 1543, 1447, 1431, 1382, 1352, 1319, 1157, 1100, 992, 942, 845, 784, 772. HRMS (ESI+): m/z [M+H]+ calcd. for C27H26ClF3N5O+, 528.1773; found, 528.1762 (error 1.98 ppm).
(6-(4-methoxybenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (26).
Yield: 90%. White solid. M.P. 53.7-55.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.38 (s, 1H), 7.39-7.44 (m, 1H), 7.21-7.24 (m, 2H), 7.03-7.08 (m, 3H), 6.82-6.85 (m, 2H), 4.08 (s, 2H), 3.74 (s, 4H), 3.69 (s, 3H), 3.34 (s, 4H), 2.78 (s, 3H), 2.45 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.7, 160.7, 157.8, 151.1, 145.4, 144.5, 142.7, 130.1, 130.0, 129.9 (q, J = 30.8 Hz), 128.8, 124.4 (q, J = 270.8 Hz), 119.0, 118.7, 114.9 (q, J = 3.7 Hz), 114.1, 111.3 (q, J = 3.7 Hz), 104.6, 55.0, 48.1 (br), 31.8, 23.7, 13.7. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −61.1. IR νmax (neat): 1613, 1583, 1510, 1445, 1383, 1312, 1245, 1113, 1025, 990, 839, 790, 699. HRMS (ESI+): m/z [M+H]+ calcd. for C28H29F3N5O2+, 524.2268; found, 524.2264 (error 0.73 ppm).
(5,7-dimethyl-6-(3-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (27).
Yield: 95%. White solid. M.P. 62.5-65.0 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.39 (s, 1H), 7.39-7.45 (m, 1H), 7.14-7.25 (m, 3H), 7.07-7.09 (m, 1H), 6.97-7.02 (m, 2H), 6.89-6.92 (m, 1H), 4.13 (s, 2H), 3.74 (s, 4H), 3.34 (s, 4H), 2.78 (s, 3H), 2.45 (s, 3H), 2.24 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.6, 160.7, 151.1, 145.3, 144.7, 142.7, 138.3, 137.8, 130.0, 129.9 (q, J = 30.8 Hz), 128.5, 128.4, 127.0, 124.8, 124.3 (q, J = 270.6 Hz), 119.0, 118.3, 114.8 (q, J = 3.4 Hz), 111.3 (q, J = 3.5 Hz), 104.6, 48.1 (br), 32.6, 23.8, 21.0, 13.7. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −61.1. IR νmax (neat): 2925, 1613, 1550, 1446, 1383, 1315, 1160, 1114, 1025, 990, 838, 791, 771, 734, 702. HRMS (ESI+): m/z [M+Na]+ calcd. for C28H28F3N5ONa+, 530.2138; found, 530.2135 (error 0.59 ppm).
(5,7-dimethyl-6-(2-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (28).
Yield: 84%. White solid. M.P. 91.1- 93.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.40 (s, 1H), 7.40-7.45 (m, 1H), 7.23-7.26 (m, 3H), 7.07-7.14 (m, 2H), 6.98-7.03 (m, 1H), 6.55-6.57 (m, 1H), 4.06 (s, 2H), 3.77 (s, 4H), 3.38 (s ,4H), 2.69 (s, 3H), 2.44 (s, 3H), 2.39 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.7, 160.8, 151.1, 145.4, 144.9, 142.9, 136.3, 136.1, 130.0, 129.9 (q, J = 30.8 Hz), 126.2, 126.2, 125.8, 124.4 (q, J = 270.9 Hz), 119.0, 117.7, 114.8 (q, J = 3.6 Hz), 111.3 (q, J = 3.6 Hz), 104.7, 48.0 (br), 30.0, 23.4, 19.4, 13.5. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −61.1. IR νmax (neat): 2914, 1615, 1548, 1432, 1352, 1318, 1307, 1159, 1117, 1027, 991, 944, 840, 765, 696. HRMS (ESI+): m/z [M+Na]+ calcd. for C28H28F3N5ONa+, 530.2138; found, 530.2130 (error 1.54 ppm).
(6-(4-isopropylbenzyl)-5,7-dimethylpyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (29).
Yield: 65%. White solid. M.P. 176.4-177.9 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.38 (s, 1H), 7.40-7.45 (m, 1H), 7.21-7.25 (m, 2H), 7.14-7.17 (m, 2H), 7.04-7.09 (m, 3H), 4.14 (s, 2H), 3.74 (s, 4H), 3.34 (s, 4H), 2.79-2.88 (m, 4H), 2.46 (s, 3H), 1.15 (d, J = 6.8 Hz, 6H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.6, 160.7, 151.0, 146.3, 145.3, 144.5, 142.7, 135.6, 130.0, 129.8 (q, J = 31.1 Hz), 127.7, 126.5, 124.3 (q, J = 270.8 Hz), 118.9, 118.4, 114.8 (q, J = 3.7 Hz), 111.2 (q, J = 3.7 Hz), 104.6, 48.0 (br), 32.9, 32.2, 23.8, 13.7. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −61.1. IR νmax (neat): 2959, 1614, 1541, 1447, 1428, 1376, 1351, 1235, 1156, 1118, 1026, 992, 846, 774, 691. HRMS (ESI+): m/z [M+H]+ calcd. for C30H33F3N5O+, 536.2632; found, 536.2631 (error 0.13 ppm).
(5,7-dimethyl-6-(4-(trifluoromethyl)benzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (30).
Yield: 60%. White solid. M.P. 202.6- 203.6 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.40 (s, 1H), 7.63-7.66 (m, 2H), 7.38-7.45 (m, 3H), 7.21-7.26 (m, 2H), 7.07-7.10 (m, 1H), 4.31 (s, 2H), 3.75 (s, 4H), 3.35 (s, 4H), 2.80 (s, 3H), 2.45 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.5, 160.5, 151.0, 145.4, 145.0, 143.5, 142.8, 129.9, 129.8 (q, J = 30.8 Hz), 128.7, 127.0 (q, J = 31.5 Hz), 125.3 (q, J = 3.6 Hz), 124.3 (q, J = 270.7 Hz), 124.2 (q, J = 270.3 Hz), 118.9, 117.4, 114.8 (q, J = 3.7 Hz), 111.2 (q, J = 3.7 Hz), 104.7, 48.0 (br), 32.5, 23.7, 13.7. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −60.8, −61.1. IR νmax (neat): 2816, 1609, 1447, 1432, 1383, 1319, 1159, 1111, 1066, 1018, 993, 943, 849, 788, 691. HRMS (ESI+): m/z [M+Na]+ calcd. for C28H25F6N5ONa+, 584.1856; found, 584.1851 (error 0.77 ppm).
(6-ethyl-5,7-dimethylpyrazolo[1,5-a]pyrimidin-3-yl)(4-phenylpiperazin-1-yl)methanone (31).
Yield: 73%. White solid. M.P. 143.0-144.4 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.33 (s, 1H), 7.19-7.25 (m, 2H), 6.95-6.98 (m, 2H), 6.77-6.82 (m, 1H), 3.72 (brs, 4H), 3.22 (brs, 4H), 2.69-2.74 (m, 5H), 2.60 (s, 3H), 1.13 (t, J = 7.3 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.6, 160.0, 150.9, 145.1, 143.3, 142.4, 129.0, 121.4, 119.2, 115.8, 104.4, 48.8 (br), 23.3, 20.6, 13.6, 12.9. IR νmax (neat): 2966, 2923, 2864, 1617, 1596, 1542, 1429, 1370, 1227, 1191, 837, 754, 689. HRMS (ESI+): m/z [2M+Na]+ calcd. for C42H50N10O2Na+, 749.4010; found, 749.4010 (error 0 ppm).
(5,7-dimethyl-6-phenylpyrazolo[1,5-a]pyrimidin-3-yl)(4-phenylpiperazin-1-yl)methanone (32).
Yield: 84%. White solid. M.P. 97.0-98.5 °C. 1H NMR (500 MHz, DMSO-d6) δ (ppm): 8.43 (s, 1H), 7.53-7.56 (m, 2H), 7.48-7.50 (m, 1H), 7.38-7.40 (m, 2H), 7.21-7.24 (m, 2H), 6.96-6.98 (m, 2H), 6.80 (t, J = 7.2 Hz, 1H), 3.77 (s, 4H), 3.24-3.26 (m, 4H), 2.48 (s, 3H), 2.29 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 162.4, 159.2, 150.9, 145.7, 143.8, 143.0, 134.9, 134.8, 129.9, 128.9, 128.8, 128.2, 122.4, 119.2, 115.8, 104.8, 48.8, 24.6, 14.8. IR νmax (neat): 3056, 2920, 2850, 1722, 1614, 1597, 1494, 1385, 1228, 839, 757, 704, 692. HRMS (ESI+): m/z [2M+Na]+ calcd. for C50H50N10O2Na+, 845.4010; found, 845.4010 (error 0 ppm).
(5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (33).
Yield: 92%. White solid. M.P. 76.5-78.1 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.39 (s, 1H), 7.50 (d, J = 8.6 Hz, 2H), 7.00-7.10 (m, 6H), 4.11 (s, 2H), 3.74 (s, 4H), 3.40 (s, 4H), 2.78 (s, 3H), 2.45 (s, 3H), 2.24 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.6, 160.6, 153.0, 145.3, 144.5, 142.7, 135.3, 135.2, 129.2, 127.7, 126.1 (q, J = 3.5 Hz), 124.8 (q, J = 268.7 Hz), 118.4, 118.0 (q, J = 31.8 Hz), 114.3, 104.6, 47.3 (br), 32.2, 23.7, 20.5, 13.6. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −59.5. IR νmax (neat): 2920, 2855, 1613, 1547, 1522, 1428, 1385, 1326, 1230, 1193, 1104, 1069, 988, 824. HRMS (ESI+): m/z [M+H]+ calcd. for C28H29F3N5O+, 508.2319; found, 508.2302 (error 3.29 ppm).
(5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(2-(trifluoromethyl)phenyl)piperazin-1-yl)methanone (34).
Yield: 91%. White solid. M.P. 171.4-173.4 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.38 (s, 1H), 7.58-7.69 (m, 3H), 7.33-7.38 (m, 1H), 7.08 (d, J = 7.7 Hz, 2H), 7.01 (d, J = 7.7 Hz, 2H), 4.12 (s, 2H), 3.72 (s, 4H), 2.93 (s ,4H), 2.77 (s, 3H), 2.46 (s, 3H), 2.24 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.5, 160.6, 151.9, 145.2, 144.5, 142.7, 135.3, 135.2, 133.6, 129.2, 127.7, 126.8 (q, J = 5.2 Hz), 125.7 (q, J = 27.8 Hz), 125.4, 124.7, 124.0 (q, J = 271.6 Hz), 118.4, 104.7, 53.4 (br), 32.2, 23.7, 20.5, 13.6. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −58.7. IR νmax (neat): 2916, 2810, 1620, 1548, 1514, 1481, 1430, 1377, 1312, 1120, 1107, 1036, 1025, 988, 771. HRMS (ESI+): m/z [M+H]+ calcd. for C28H29F3N5O+, 508.2319; found, 508.2304 (error 2.89 ppm).
(4-(3-(difluoromethyl)phenyl)piperazin-1-yl)(5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)methanone (35).
Yield: 89%. White solid. M.P. 82.3-84.3 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.38 (s, 1H), 7.32-7.37 (m, 1H), 7.07-7.13 (m, 4H), 6.96-7.03 (m, 3H), 6.92 (t, J = 56.0 Hz, 1H), 4.12 (s, 2H), 3.74 (s, 4H), 3.30 (s, 4H), 2.78 (s, 3H), 2.45 (s, 3H), 2.24 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.6, 160.7, 150.9, 145.3, 144.5, 142.7, 135.3, 135.2, 134.8 (t, J = 21.7 Hz), 129.6, 129.2, 127.7, 118.4, 118.3, 117.8 (br), 115.7 (t, J = 5.7 Hz), 115.2 (t, J = 234.7 Hz), 112.5 (t, J = 6.3 Hz), 112.1, 104.6, 48.3 (br), 32.2, 23.7, 20.5, 13.6. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −109.0. IR νmax (neat): 2921, 1614, 1548, 1480, 1432, 1377, 1236, 1192, 1026, 992, 840, 792, 774, 699. HRMS (ESI+): m/z [M+Na]+ calcd. for C28H29F2N5ONa+, 512.2232; found, 512.2232 (error 0 ppm).
3-(4-(5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidine-3-carbonyl)piperazin-1-yl)benzo nitrile (36).
Yield: 94%. White solid. M.P. 83.8-85.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.38 (s, 1H), 7.37-7.42 (m, 2H), 7.29-7.31 (m, 1H), 7.16-7.19 (m, 1H), 7.09 (d, J = 7.8 Hz, 2H), 7.02 (d, J = 7.8 Hz, 2H), 4.13 (s, 2H), 3.72 (s, 4H), 3.34 (s, 4H), 2.78 (s, 3H), 2.45 (s, 3H), 2.25 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.6, 160.7, 150.8, 145.3, 144.6, 142.7, 135.3, 135.2, 130.2, 129.2, 127.7, 121.8, 119.9, 119.2, 118.4, 117.9, 112.0, 104.6, 47.7 (br), 32.2, 23.8, 20.5, 13.7. IR νmax (neat): 2921, 2227, 1615, 1595, 1547, 1430, 1376, 1236, 1193, 1028, 991, 840, 772. HRMS (ESI+): m/z [M+H]+ calcd. for C28H29N6O+, 465.2397; found, 465.2401 (error −0.78 ppm).
(5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-fluorophenyl)piperazin-1-yl)methanone (37).
Yield: 96%. White solid. M.P. 62.0-63.4 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.38 (s, 1H), 7.17-7.25 (m, 1H), 7.09 (d, J = 7.8 Hz, 2H), 7.02 (d, J = 7.8 Hz, 2H), 6.74-6.79 (m, 2H), 6.53-6.58 (m, 1H), 4.12 (s, 2H), 3.72 (s, 4H), 3.28 (s, 4H), 2.78 (s, 3H), 2.45 (s, 3H), 2.25 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.2 (d, J = 238.9 Hz), 162.6, 160.7, 152.5 (d, J = 10.0 Hz), 145.3, 144.5, 142.7, 135.3, 135.2, 130.3 (d, J = 10.0 Hz), 129.2, 127.7, 118.4, 111.1, 104.9 (d, J = 21.2 Hz), 104.6, 102.0 (d, J = 25.0 Hz), 48.1 (br), 32.2, 23.7, 20.5, 13.6. 19F NMR (282 MHz, DMSO-d6) δ (ppm): −112.3. IR νmax (neat): 2919, 1668, 1611, 1582, 1547, 1494, 1434, 1385, 1243, 1160, 992, 968, 834, 772. HRMS (ESI+): m/z [2M+Na]+ calcd. for C54H56F2N10O2Na+, 937.4448; found, 937.4448 (error 0 ppm).
(4-(3-chlorophenyl)piperazin-1-yl)(5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)methanone (38).
Yield: 70%. White solid. M.P. 70.6-72.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.37 (s, 1H), 7.18-7.23 (m, 1H), 7.08 (d, J = 7.8 Hz, 2H), 7.01 (d, J = 7.8 Hz, 2H), 6.89-6.96 (m, 2H), 6.77-6.80 (m, 1H), 4.11 (s, 2H), 3.72 (s, 4H), 3.28 (s, 4H), 2.78 (s, 3H), 2.45 (s, 3H), 2.24 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.5, 160.6, 152.0, 145.3, 144.5, 142.6, 135.3, 135.2, 133.8, 130.4, 129.2, 127.7, 118.4, 118.3, 114.9, 113.9, 104.6, 48.0 (br), 32.2, 23.7, 20.5, 13.6. IR νmax (neat): 2916, 1618, 1589, 1550, 1479, 1429, 1383, 1192, 1025, 989, 774. HRMS (ESI+): m/z [M+H]+calcd. for C27H29ClN5O+, 474.2055; found, 474.2058 (error −0.60 ppm).
(5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-methoxyphenyl)piperazin-1-yl)methanone (39).
Yield: 85%. White solid. M.P. 60.1-61.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.38 (s, 1H), 7.07-7.13 (m, 3H), 7.01 (d, J = 7.8 Hz, 2H), 6.48-6.54 (m, 2H), 6.36-6.39 (m, 1H), 4.10 (s, 2H), 3.70 (m, 7H), 3.22 (s, 4H), 2.77 (s, 3H), 2.44 (s, 3H), 2.23 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.5, 160.6, 160.2, 152.2, 145.3, 144.5, 142.6, 135.3, 135.2, 129.6, 129.2, 127.7, 118.4, 108.3, 104.7, 104.5, 101.9, 54.8, 48.7 (br), 32.2, 23.7, 20.5, 13.6. IR νmax (neat): 2920, 1615, 1594, 1549, 1481, 1445, 1382, 1256, 1191, 1163, 1024, 991, 840, 774. HRMS (ESI+): m/z [M+Na]+ calcd. for C28H31N5O2Na+, 492.2370; found, 492.2366 (error 0.80 ppm).
(5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(m-tolyl)piperazin-1-yl)methano ne (40).
Yield: 79%. White solid. M.P. 78.9-80.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.37 (s, 1H), 7.07-7.11 (m, 3H), 7.02 (d, J = 7.8 Hz, 2H), 6.74-6.78 (m, 2H), 6.60-6.63 (m, 1H), 4.12 (s, 2H), 3.71 (s, 4H), 3.20 (s ,4H), 2.78 (s, 3H), 2.45 (s, 3H), 2.24-2.25 (m, 6H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.5, 160.6, 150.9, 145.3, 144.5, 142.7, 138.0, 135.3, 135.2, 129.2, 128.8, 127.7, 120.0, 118.4, 116.5, 113.0, 104.7, 48.8 (br), 32.2, 23.8, 21.4, 20.5, 13.7. IR νmax (neat): 2920, 1669, 1615, 1600, 1547, 1433, 1385, 1241, 1193, 991, 836, 772, 693. HRMS (ESI+): m/z [M+Na]+ calcd. for C28H31N5ONa+, 476.2421; found, 476.2408 (error 2.69 ppm).
(5,7-dimethyl-6-(4-methylbenzyl)pyrazolo[1,5-a]pyrimidin-3-yl)(4-(3-isopropylphenyl)piperazin-1-yl)methanone (41).
Yield: 75%. White solid. M.P. 78.9-80.5 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.38 (s, 1H), 7.08-7.15 (m, 3H), 7.02 (d, J = 7.7 Hz, 2H), 6.83 (s, 1H), 6.74-6.77 (m, 1H), 6.67-6.70 (m, 1H), 4.12 (s, 2H), 3.72 (s, 4H), 3.20 (s 4H), 2.78-2.84 (m, 4H), 2.45 (s, 3H), 2.24 (s, 3H), 1.16 (d, J = 6.8 Hz, 6H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.5, 160.6, 151.0, 149.2, 145.3, 144.5, 142.7, 135.3, 135.2, 129.2, 128.8, 127.7, 118.4, 117.3, 114.3, 113.4, 104.7, 49.0 (br), 33.7, 32.2, 23.9, 23.7, 20.5, 13.7. IR νmax (neat): 2958, 1668, 1615, 1599, 1548, 1434, 1385, 1237, 1196, 993, 835, 776, 700. HRMS (ESI+): m/z [M+Na]+ calcd. for C30H35N5ONa+, 504.2734; found, 504.2732 (error 0.36 ppm).
6-benzyl-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxamide (42).
Yield: 60%. White solid. M.P. 245.5-248.0 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.46 (s, 1H), 7.57 (s, 1H), 7.42 (s, 1H), 7.26-7.31 (m, 2H), 7.18-7.23 (m, 1H), 7.12-7.15 (m, 2H), 4.20 (s, 2H), 2.79 (s, 3H), 2.50 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 163.0, 161.5, 145.5, 144.8, 144.0, 138.3, 128.7, 127.9, 126.4, 118.7, 104.4, 32.6, 23.7, 13.7. IR νmax (neat): 3396, 3314, 3272, 3155, 2940, 1651, 1614, 1541, 1524, 1485, 1267, 1196, 797, 704, 635. HRMS (ESI+): m/z [2M+H]+ calcd. for C32H33N8O2+, 561.2721; found, 561.2708 (error 2.31 ppm).
6-benzyl-N-cyclohexyl-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carboxamide (43).
Yield:57%. White solid. M.P. 180.1-181.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.46 (s, 1H), 8.07 (d, J = 7.7 Hz, 1H), 7.26-7.31 (m, 2H), 7.17-7.22 (m, 1H), 7.12-7.14 (m, 2H), 4.19 (s, 2H), 3.86 (m, 1H), 2.79 (s, 3H), 2.50 (s, 3H), 1.84-1.87 (m, 2H), 1.66 (m, 2H), 1.52 (m, 1H), 1.27-1.40 (m, 5H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 161.3, 160.4, 145.7, 144.4, 143.7, 138.2, 128.6, 127.8, 126.4, 118.7, 104.3, 46.4, 32.6, 32.4, 25.2, 24.0, 13.7. IR νmax (neat): 3314, 2929, 2851, 1643, 1612, 1548, 1517, 1427, 1194, 778, 743, 703, 645. HRMS (ESI+): m/z [2M+H]+ calcd. for C44H53N8O2+, 725.4286; found, 725.4286 (error 0 ppm).
6-benzyl-5,7-dimethyl-N-phenylpyrazolo[1,5-a]pyrimidine-3-carboxamide (44).
Yield: 75%. White solid. M.P. 211.3-212.6 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 10.19 (s, 1H), 8.64 (s, 1H), 7.73-7.76 (m, 2H), 7.34-7.39 (m, 2H), 7.28-7.31 (m, 2H), 7.22-7.24 (m, 1H), 7.16-7.19 (m, 2H), 7.09 (m, 1H), 4.25 (s, 2H), 2.85 (s, 3H), 2.62 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.0, 159.8, 146.1, 144.9, 143.7, 138.8, 138.2, 129.0, 128.7, 127.9, 126.4, 123.3, 119.3, 119.1, 104.0, 32.6, 23.9, 13.8. IR νmax (neat): 3297, 3256, 3197, 3146, 1779, 1662, 1600, 1543, 1499, 1481, 1381, 1312, 1257, 1196, 744, 706. HRMS (ESI+): m/z [M+H]+ calcd. for C22H21N4O+, 357.1710; found, 357.1713 (error −0.88 ppm).
(6-benzyl-5,7-dimethylpyrazolo[1,5-a]pyrimidin-3-yl)(piperidin-1-yl)methanone (45).
Yield: 96%. White solid. M.P. 69.2-71.0 °C. 1H NMR (500 MHz, DMSO-d6) δ (ppm): 8.30 (s, 1H), 7.27-7.30 (m, 2H), 7.18-7.21 (m, 1H), 7.12-7.14 (m, 2H), 4.17 (s, 2H), 3.53 (brs, 4H), 2.77 (s, 3H), 2.44 (s, 3H), 1.59-1.61 (m, 2H), 1.55-1.56 (m, 4H). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 162.3, 160.3, 144.9, 144.4, 142.6, 138.5, 128.6, 127.8, 126.3, 118.0, 105.3, 48.2 (br), 42.6 (br), 32.6, 25.8 (br), 24.1, 23.7, 13.7. IR νmax (neat): 2933, 2853, 1733, 1674, 1614, 1547, 1434, 1386, 1252, 982, 835, 770, 734, 703, 655. HRMS (ESI+): m/z [2M+Na]+ calcd. for C42H48N8O2Na+, 719.3792; found, 719.3792 (error 0 ppm).
tert-Butyl 4-(6-benzyl-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carbonyl)piperazine-1-carboxylate (46).
Yield: 86%. White solid. M.P. 186.3-188.8 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.35 (s, 1H); 7.27-7.31 (m, 2H), 7.21-7.23 (m, 1H), 7.13-7.15 (m, 2H), 4.18 (s, 2H), 3.55(s, 4H), 3.44(s, 4H), 2.78 (s, 3H), 2.46 (s, 3H), 1.41 (s, 9H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.7, 160.7, 153.8, 145.4, 144.7, 142.7, 138.4, 128.7, 127.9, 126.3, 118.3, 104.6, 79.1, 43.8, 32.6, 28.0, 23.8, 13.7. IR νmax (neat): 3100, 2990, 2851, 1697, 1609, 1541, 1418, 1364, 1237, 1166, 837, 772, 742, 708, 657. HRMS (ESI): m/z [M+H]+ calcd. for C25H32N5O3+, 450.2500; found, 450.2508 (error −1.85 ppm).
(6-benzyl-5,7-dimethylpyrazolo[1,5-a]pyrimidin-3-yl)(piperazin-1-yl)methanone (47).
tert-Butyl 4-(6-benzyl-5,7-dimethylpyrazolo[1,5-a]pyrimidine-3-carbonyl)piperazine-1-carboxylate 46 (600 mg, 1.33 mmol, 1.0 eq.) was dissolved in 6 mL of 4 M HCl in EtOAc. The solution was stirred at room temperature for 4h, and then the solvent was removed in vacuo. The residue was partitioned between 30 mL DCM and 30 mL saturated sodium bicarbonate. The aqueous layer was extracted with DCM (20 mL×2). The combined organic layers were dried over anhydrous Na2SO4 and concentrated in vacuo to give compound 47 (173 mg, 0.49 mmol). Yield: 37%. White solid. M.P. 53.7-55.4 °C. 1H NMR (300 MHz, DMSO-d6) δ (ppm): 8.32 (s, 1H), 7.26-7.30 (m, 2H), 7.17-7.22 (m, 1H), 7.12-7.14 (m, 2H), 4.16 (s, 2H), 3.49 (s, 4H), 2.88-2.98 (m, 1H), 2.77 (s, 3H), 2.74 (s, 4H), 2.44 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ (ppm): 162.4, 160.3, 145.0, 144.5, 142.6, 138.4, 128.6, 127.8, 126.3, 118.0, 105.0, 45.9, 32.6, 23.7, 13.6. IR νmax (neat): 3491, 3311, 2918, 2852, 1732, 1614, 1547, 1427, 1386, 1230, 1192, 1027, 988, 769, 735, 705. HRMS (ESI+): m/z [M+H]+ calcd. for C20H24N5O+, 350.1975; found, 350.1981 (error −1.61 ppm).
(6-benzyl-5,7-dimethylpyrazolo[1,5-a]pyrimidin-3-yl)(4-(prop-2-yn-1-yl)piperazin-1-yl)methanone (48).
(6-benzyl-5,7-dimethylpyrazolo[1,5-a]pyrimidin-3-yl)(piperazin-1-yl)methanone 47 (240 mg, 0.69 mmol, 1.0 eq.) was dissolved in anhydrous DCM (10 mL), and anhydrous K2CO3 (477 mg, 3.45 mmol, 5.0 eq.) was added. The mixture was vigorously stirred at room temperature, and neat propargyl bromide (204 mg, 1.72 mmol, 2.5 eq.) was added. The reaction was stirred at room temperature overnight, K2CO3 was filtered, and the filtrate was concentrated in vacuo. The residue was purified by column chromatography (silica gel, Dichloromethane/Methanol, 200:1 to 30:1) to give compound 48. (36 mg, 0.09 mmol) Yield: 13%. White solid. M.P. 177.2-179.3 °C. 1H NMR (500 MHz, DMSO-d6) δ (ppm): 8.34 (s, 1H), 7.29 (m, 2H), 7.20 (m, 1H), 7.13-7.14 (m, 2H), 4.18 (s, 2H), 3.59 (brs, 4H), 3.32 (d, J = 2.2 Hz, 2H), 3.18 (t, J = 2.2 Hz, 1H), 2.77 (s, 3H), 2.52-2.54 (m, 4H), 2.45 (s, 3H). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 162.5, 160.6, 145.2, 144.6, 142.7, 138.5, 128.7, 127.9, 126.4, 118.2, 104.8, 79.1, 76.0, 51.6, 47.1, 46.0, 41.8, 32.6, 23.8, 13.7. IR νmax (neat): 3290, 3023, 2993, 2922, 2856, 2828, 1614, 1552, 1451, 1430, 1376, 1234, 1193, 1138, 1000, 888, 709. HRMS (ESI+): m/z [2M+H]+ calcd. for C46H51N10O2+, 775.4191; found, 775.4191 (error 0 ppm).
4.2. Docking-Based Virtual Screening
The NCI compound library containing approximately 260,000 compounds was screened through the MTiOpen screen automated virtual screening platform using the integrated Vina AutoDock program against RUVBL1/2 complex (PDB code: 5oaf) [38, 39]. The duplicate compounds were removed using Mona [40]. The 3D structure of all the compounds were generated and the energy minimization was performed by Frog2 [41]. The settings for compound library filters were as follows: the number of hydrogen bond acceptors < 10, the number of hydrogen bonds donors < 5, LogP < 5, molecular weight < 500, the number of rotatable bonds < 10, the topological polar surface area <140 [42]. The Grid center coordinates were 14.5666, 189.849, and 166.659. The sizes of the search space [Å] was 50 × 50 × 50. The top 3,000 results were visualized, and 80 compounds were selected to evaluate the inhibition activity against RUVBL1/2 complex.
4.3. Molecular cloning and Sequencing
The cDNA sequences of RUVBL1 and RUVBL2 were amplified using Platinum SuperFi PCR Master Mix (Thermofisher, Cat. No. 12358-010). PCR product was purified after electrophoresis through 0.8% Agarose and extracted using QIAquick Gel Extraction kit (QIAGEN, Cat. No. 28115). Both RUVBL1 and RUVBL2 were inserted into pCOLA-duet vector by Gibson assembly. Molecular cloning was performed using a NEB PCR Cloning Kit (Biolabs, NEB #E1202) according to the manufacturer’s protocol. After overnight incubation, 5 clones were picked and plasmids were purified using a Plasmid miniprep kit (Bioland, Cat. No. PD01-01). Purified plasmids were sequenced by Sanger sequencing. PCR and sequencing primers are listed in Tables S4 and S5.
4.4. Expression and Purification of Recombinant Protein
Full-length recombinant His6x-tagged human RUVBL1 and RUVBL2 were expressed in E. coli Rosetta cells. The Rosetta cells were suspended in lysis buffer composed of 20 mM Tris, pH 8.0, 200 mM NaCl, 4 mM MgCl2, 20 mM imidazole, β-mercaptoethanol, 50 mL/pill protease inhibitor, and 5% glycerol. The cell suspension was sonicated for three rounds at 65, 70 and 70 amp, respectively. The mixture was then centrifuged for 1.5 hours at 15000 x g. The resulting supernatant was loaded onto a Ni-NTA column and eluted with a linear gradient of imidazole (0 min, 20 mM Tris pH 8.0, 300 mM NaCl, 4 mM MgCl2, 20 mM Imidazole, 1 mM DTT, and 5% glycerol; 50 min, 20 mM Tris pH 8.0, 300 mM NaCl, 4 mM MgCl2, 400 mM Imidazole, 1 mM DTT, and 5% glycerol; 60 min, 20 mM Tris pH 8.0, 300 mM NaCl, 4 mM MgCl2, 400 mM Imidazole, 1 mM DTT, and 5% glycerol). SDS-Page was employed to probe the fractions containing RUVBL1/2 complex. The RUVBL1/2 solution was condensed and further purified by size-exclusion chromatography in 20 mM Tris pH 7.5, 300 mM sodium chloride, 1 mM magnesium chloride, 1 mM DTT, and 10% glycerol using a Superose 6 Increase 10/300 GL column (GE Healthcare), and stored in aliquots at −80 °C with glycerol.
4.5. ATPase Assay
The purified protein was assayed in 50 μL ATPase assay buffer (50 mM Tris pH 7.4, 20 mM MgCl2, 0.1% Glycerol, and 0.01% Triton X-100) containing 200 μM ATP. After 70 min incubation at 37 °C, 50 μL Biomol Green reagent (Enzo Life Sciences) was added to quench the reaction, and the absorbance at 635 nm was measured using a BioTek Synergy Neo 2 plate reader. The eight-dose titrations (0, 2.00, 6.58, 9.88, 14.81, 22.22, 33.33, 50.00 μM) were performed of the compound to the reaction to determine the IC50 values of the compounds. In this assay, the concentration of RUVBL1/2, RUVBL, and RUVBL2 were 0.05 μM, 1.0 μM, and 3.0 μM, respectively. The results were calculated from six replicates using GraphPad Prism 7.0.
4.6. Antiproliferative assay
Cell viability was measured using CellTiter Glo® Luminescent Cell Viability Assay (Promega) according to the manufacturer’s procedure. RMPI1640 or DMEM containing 5% FBS and 1% Penicillin Streptomycin (Thermo Fisher) was used as cell viability assay medium. To find the linear relationship between the relative luminescence unit and the number of viable cells, a standard curve for each cell line was generated. Generally, 30 μL of cell suspension was plated in 384-well white plates (Greiner) with serial 2-fold dilution (from 30000 to 284 cells per well). Twenty-four hours after seeding, 8 μL of assay media containing 5% DMSO was added into each well and plates were incubated for an additional 48 h at 37 °C in a 5% CO2 incubator. To test the anti-proliferative activity of RUVBL1/2 complex inhibitors, cells were seeded at 750 to 3000 cells per well according to the linear range determined from the standard curve of each cell line. Twenty-four hours after seeding, cells were treated with the compounds (three-fold dilution, eight concentrations). After 48 hours of treatment, cell viability was measured using CellTiter Glo and IC50 values were calculated using the percentage of growth of treated cells versus the DMSO control.
4.7. Proteomics
HCT116 cells were treated with DMSO, 15 μM or 30 μM tested compound for 24 h, and pellets were harvested. The LC-MS samples were prepared by following the instructions of Thermo EasyPep Mini MS Sample Prep Kit (REF A4006) and peptide concentration was tested through Pierce Quantitative Fluorometric Peptide Assay (cat# 23290).
The LC-MS/MS experiments were performed using an EASY-nLC 1000 (ThermoFisher Scientific, San Jose, CA) connected to an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The sample (1 μg) in 0.1% FA solution was loaded onto an Aurora UHPLC Column (25 cm x 75 μm, 1.6 μm C18, AUR2-25075C18A, Ion Opticks) and separated over 136 min at a flow rate of 0.35 μL/min with the following gradient: 2-6% Solvent B (7.5 min), 6-25% B (82.5 min), 25-40% B (30 min), 40-98% B (1 min), and 98% B (15 min). Solvent A consisted of 97.9% H2O, 2% ACN, and 0.1% formic acid, and solvent B consisted of 19.9% H2O, 80% ACN, and 0.1% formic acid. An MS1 scan was acquired in the Orbitrap at 120,000 resolution with a scan range of 350-1500 m/z. The AGC target was 4x105, and the maximum injection time was 50 mins. Dynamic exclusion was set to exclude features after 1 time for 60 s with a 10-ppm mass tolerance. Higher-energy collisional dissociation (HCD) fragmentation was performed with 35% collision energy after quadruple isolation of features using a 1.6 m/z isolation window, 5×104 AGC target, and 35 ms maximum injection time. MS2 scans were then also acquired by the Orbitrap with 50,000 resolution. Ion source settings were as follows: ion source type, NSI; spray voltage, 2400 V; ion transfer tube temperature, 275 °C. System control and data collection were performed using Xcalibur software.
The proteomic analysis was performed through Proteome Discoverer 2.4 (Thermo Scientific) using the Uniprot human database and the Byonic search algorithm (Protein Metrics). Percolator FDRs were set at 0.001 (strict) and 0.01 (relaxed). Peptide FDRs were set at 0.001 (strict) and 0.01 (relaxed), with medium confidence and a minimum peptide length of 6. Normalization was performed on the total peptide amount. The limma analysis was performed using R studio following the user guide [43]. The volcano and heatmap figures were plotted by GraphPad Prism 7.0. The Venn diagram was plotted using an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
4.8. GO and KEGG pathway analysis of the differentially expressed proteins
The DAVID 6.8 database (https://david.ncifcrf.gov/) was employed to perform GO functional and KEGG pathway analysis of the integrated differentially expressed proteins [44]. The GO functional analysis of the integrated differentially expressed proteins involves three parts: biological process, cellular component and molecular function. The bubble graph was plotted by http://www.bioinformatics.com.cn, an online platform for data analysis and visualization.
Supplementary Material
Figure 4.
Docking study of compound 18 with RUVBL1/2 (PDB code 5OAF). Compound 18 is presented as a cyan ball and stick model. Active residues are presented as gray sticks. Hydrogen bond, cation-π interaction, and halogen bond are presented as dashed yellow, magenta, and green lines, respectively.
Acknowledgments
This work was supported in part by the National Institute of Child Health and Human Development R01 HD086596 and The Cultivation Fund of School of Pharmaceutical Sciences, Capital Medical University (2021).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors have no conflicts of interest to declare.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/xx.xxxx/xxxxxxxxxxxx.
References
- [1].Jha S, Dutta A, RVB1/RVB2: running rings around molecular biology, Mol. Cell, 34 (2009) 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE, Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly, Cell, 132 (2008) 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Grigoletto A, Lestienne P, Rosenbaum J, The multifaceted proteins Reptin and Pontin as major players in cancer, Biochim. Biophys. Acta, 1815 (2011) 147–157. [DOI] [PubMed] [Google Scholar]
- [4].Jha S, Gupta A, Dar A, Dutta A, RVBs are required for assembling a functional TIP60 complex, Mol. Cell Biol, 33 (2013) 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, Hopfner KP, Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex, Cell, 154 (2013) 1207–1219. [DOI] [PubMed] [Google Scholar]
- [6].Nguyen VQ, Ranjan A, Stengel F, Wei D, Aebersold R, Wu C, Leschziner AE, Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1, Cell, 154 (2013) 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kakihara Y, Houry WA, The R2TP complex: discovery and functions, Biochim. Biophys. Acta, 1823 (2012) 101–107. [DOI] [PubMed] [Google Scholar]
- [8].Yenerall P, Das AK, Wang S, Kollipara RK, Li LS, Villalobos P, Flaming J, Lin YF, Huffman K, Timmons BC, Gilbreath C, Sonavane R, Kinch LN, Rodriguez-Canales J, Moran C, Behrens C, Hirasawa M, Takata T, Murakami R, Iwanaga K, Chen BPC, Grishin NV, Raj GV, Wistuba II, Minna JD, Kittler R, RUVBL1/RUVBL2 ATPase Activity Drives PAQosome Maturation, DNA Replication and Radioresistance in Lung Cancer, Cell Chem. Biol, 27 (2020) 105–121. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nano N, Houry WA, Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes, Philos. Trans. R. Soc. Lond., B, Biol. Sci, 368 (2013) 20110399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Machado-Pinilla R, Liger D, Leulliot N, Meier UT, Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs, RNA, 18 (2012) 1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, Wo ZG, Kemler R, Kingston R, Wu C, Fishman M, Reptin and Pontin Antagonistically Regulate Heart Growth in Zebrafish Embryos, Cell, 111 (2002) 661–672. [DOI] [PubMed] [Google Scholar]
- [12].Lee JS, Kim Y, Kim IS, Kim B, Choi HJ, Lee JM, Shin HJ, Kim JH, Kim JY, Seo SB, Lee H, Binda O, Gozani O, Semenza GL, Kim M, Kim KI, Hwang D, Baek SH, Negative regulation of hypoxic responses via induced Reptin methylation, Mol. Cell, 39 (2010) 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee JS, Kim Y, Bhin J, Shin HJ, Nam HJ, Lee SH, Yoon JB, Binda O, Gozani O, Hwang D, Baek SH, Hypoxia-induced methylation of a pontin chromatin remodeling factor, Proc. Natl. Acad. Sci. U. S. A, 108 (2011) 13510–13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Torreira E, Jha S, Lopez-Blanco JR, Arias-Palomo E, Chacon P, Canas C, Ayora S, Dutta A, Llorca O, Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes, Structure, 16 (2008) 1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gorynia S, Bandeiras TM, Pinho FG, McVey CE, Vonrhein C, Round A, Svergun DI, Donner P, Matias PM, Carrondo MA, Structural and functional insights into a dodecameric molecular machine - the RuvBL1/RuvBL2 complex, J. Struct. Biol, 176 (2011) 279–291. [DOI] [PubMed] [Google Scholar]
- [16].Petukhov M, Dagkessamanskaja A, Bommer M, Barrett T, Tsaneva I, Yakimov A, Queval R, Shvetsov A, Khodorkovskiy M, Kas E, Grigoriev M, Large-scale conformational flexibility determines the properties of AAA+ TIP49 ATPases, Structure, 20 (2012) 1321–1331. [DOI] [PubMed] [Google Scholar]
- [17].Cheung KL, Huen J, Houry WA, Ortega J, Comparison of the multiple oligomeric structures observed for the Rvb1 and Rvb2 proteins, Biochem. Cell Biol, 88 (2010) 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lauscher JC, Loddenkemper C, Kosel L, Grone J, Buhr HJ, Huber O, Increased pontin expression in human colorectal cancer tissue, Hum. Pathol, 38 (2007) 978–985. [DOI] [PubMed] [Google Scholar]
- [19].Huber O, Menard L, Haurie V, Nicou A, Taras D, Rosenbaum J, Pontin and reptin, two related ATPases with multiple roles in cancer, Cancer Res., 68 (2008) 6873–6876. [DOI] [PubMed] [Google Scholar]
- [20].Lauscher JC, Elezkurtaj S, Dullat S, Lipka S, Grone J, Buhr HJ, Huber O, Kruschewski M, Increased Pontin expression is a potential predictor for outcome in sporadic colorectal carcinoma, Oncol. Rep, 28 (2012) 1619–1624. [DOI] [PubMed] [Google Scholar]
- [21].Haurie V, Menard L, Nicou A, Touriol C, Metzler P, Fernandez J, Taras D, Lestienne P, Balabaud C, Bioulac-Sage P, Prats H, Zucman-Rossi J, Rosenbaum J, Adenosine triphosphatase pontin is overexpressed in hepatocellular carcinoma and coregulated with reptin through a new posttranslational mechanism, Hepatology, 50 (2009) 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Grigoletto A, Neaud V, Allain-Courtois N, Lestienne P, Rosenbaum J, The ATPase activity of reptin is required for its effects on tumor cell growth and viability in hepatocellular carcinoma, Mol. Cancer Res, 11 (2013) 133–139. [DOI] [PubMed] [Google Scholar]
- [23].Menard L, Taras D, Grigoletto A, Haurie V, Nicou A, Dugot-Senant N, Costet P, Rousseau B, Rosenbaum J, In vivo silencing of Reptin blocks the progression of human hepatocellular carcinoma in xenografts and is associated with replicative senescence, J. Hepatol, 52 (2010) 681–689. [DOI] [PubMed] [Google Scholar]
- [24].Breig O, Bras S, Martinez Soria N, Osman D, Heidenreich O, Haenlin M, Waltzer L, Pontin is a critical regulator for AML1-ETO-induced leukemia, Leukemia, 28 (2014) 1271–1279. [DOI] [PubMed] [Google Scholar]
- [25].Elkaim J, Castroviejo M, Bennani D, Taouji S, Allain N, Laguerre M, Rosenbaum J, Dessolin J, Lestienne P, First identification of small-molecule inhibitors of Pontin by combining virtual screening and enzymatic assay, Biochem. J, 443 (2012) 549–559. [DOI] [PubMed] [Google Scholar]
- [26].Elkaim J, Lamblin M, Laguerre M, Rosenbaum J, Lestienne P, Eloy L, Cresteil T, Felpin FX, Dessolin J, Design, synthesis and biological evaluation of Pontin ATPase inhibitors through a molecular docking approach, Bioorg. Med. Chem. Lett, 24 (2014) 2512–2516. [DOI] [PubMed] [Google Scholar]
- [27].Ebisawa M, Suzuki T, Haginoya N, Hamada T, Murata T, Uoto K, Murakami R, Takata T, Aminopyrazolone derivative, CA2939687A1, 2017. [Google Scholar]
- [28].Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S, AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay, Sci. Signal, 3 (2010) ra27. [DOI] [PubMed] [Google Scholar]
- [29].Nano N, Ugwu F, Seraphim TV, Li T, Azer G, Isaac M, Prakesch M, Barbosa LRS, Ramos CHI, Datti A, Houry WA, Sorafenib as an Inhibitor of RUVBL2, Biomolecules, 10 (2020) 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M, Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling, Mol. Cancer Ther, 7 (2008) 3129–3140. [DOI] [PubMed] [Google Scholar]
- [31].Assimon VA, Tang Y, Vargas JD, Lee GJ, Wu ZY, Lou K, Yao B, Menon MK, Pios A, Perez KC, Madriaga A, Buchowiecki PK, Rolfe M, Shawver L, Jiao X, Le Moigne R, Zhou HJ, Anderson DJ, CB-6644 Is a Selective Inhibitor of the RUVBL1/2 Complex with Anticancer Activity, ACS Chem. Biol, 14 (2019) 236–244. [DOI] [PubMed] [Google Scholar]
- [32].Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem, 30 (2009) 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zoete V, Daina A, Bovigny C, Michielin O, SwissSimilarity: A Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening, J. Chem. Inf. Model, 56 (2016) 1399–1404. [DOI] [PubMed] [Google Scholar]
- [34].Wang R, Li X, Sun C, Yu L, Hua D, Shi C, Wang Q, Rao C, Luo W, Jiang Z, Zhou X, Yu S, The ATPase Pontin is a key cell cycle regulator by amplifying E2F1 transcription response in glioma, Cell Death Dis., 12 (2021) 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shen X, Mizuguchi G, Hamiche A, Wu C, A chromatin remodelling complex involved in transcription and DNA processing, Nature, 406 (2000) 541–544. [DOI] [PubMed] [Google Scholar]
- [36].Yenerall P, Das AK, Wang S, Kollipara RK, Li LS, Villalobos P, Flaming J, Lin YF, Huffman K, Timmons BC, Gilbreath C, Sonavane R, Kinch LN, Rodriguez-Canales J, Moran C, Behrens C, Hirasawa M, Takata T, Murakami R, Iwanaga K, Chen BPC, Grishin NV, Raj GV, Wistuba II, Minna JD, Kittler R, RUVBL1/RUVBL2 ATPase Activity Drives PAQosome Maturation, DNA Replication and Radioresistance in Lung Cancer, Cell Chem. Biol, 27 (2020) 105–121.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang G, Li S, Cheng KW, Chou TF, AAA ATPases as therapeutic targets: Structure, functions, and small-molecule inhibitors, Eur. J. Med. Chem, 219 (2021) 113446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Labbe CM, Rey J, Lagorce D, Vavrusa M, Becot J, Sperandio O, Villoutreix BO, Tuffery P, Miteva MA, MTiOpenScreen: a web server for structure-based virtual screening, Nucleic Acids Res., 43 (2015) W448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Aramayo RJ, Willhoft O, Ayala R, Bythell-Douglas R, Wigley DB, Zhang X, Cryo-EM structures of the human INO80 chromatin-remodeling complex, Nat. Struct. Mol. Biol, 25 (2018) 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hilbig M, Rarey M, MONA 2: A Light Cheminformatics Platform for Interactive Compound Library Processing, J. Chem. Inf. Model., 55 (2015) 2071–2078. [DOI] [PubMed] [Google Scholar]
- [41].Miteva MA, Guyon F, Tuffery P, Frog2: Efficient 3D conformation ensemble generator for small compounds, Nucleic Acids Res., 38 (2010) W622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD, Molecular properties that influence the oral bioavailability of drug candidates, J. Med. Chem, 45 (2002) 2615–2623. [DOI] [PubMed] [Google Scholar]
- [43].Smyth GK, Thorne NP, Wettenhall J, Limma: Linear Models for Microarray Data User's Guide. Software manual available from http://www.bioconductor.org., 2003. [Google Scholar]
- [44].Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA, Extracting biological meaning from large gene lists with DAVID, Curr. Protoc. Bioinformatics, 27 (2009) 1–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.