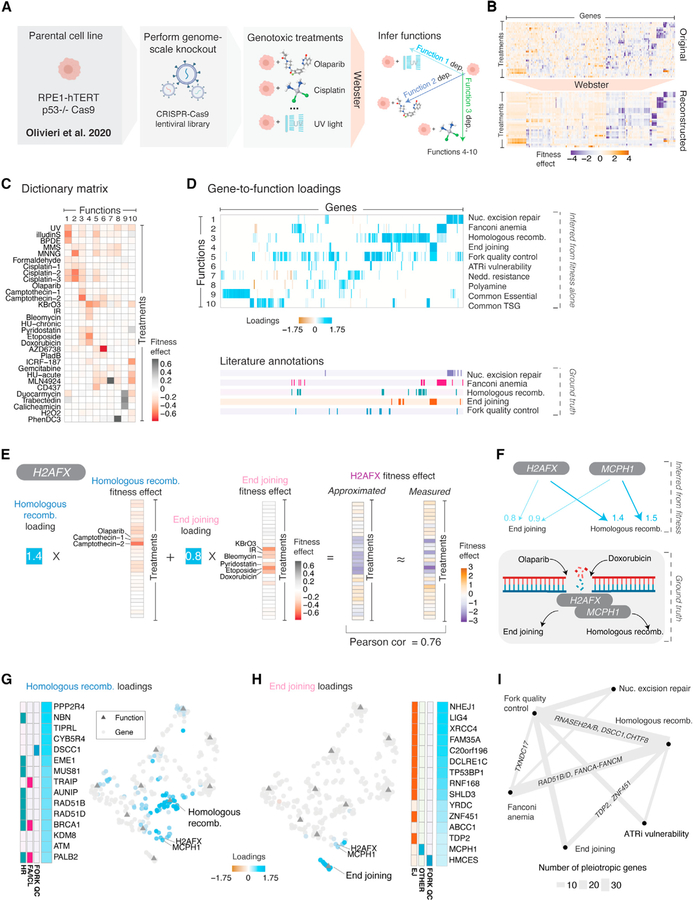
Figure 2. Pleiotropy underlies the DNA damage response to genotoxins in a human cell line.
(A) The immortalized human cell line RPE1-hTERT harboring a genome-scale CRISPR-Cas9 knockout library was subjected to 31 genotoxic stressors at a sublethal dose, resulting in a genotoxic fitness screen collection (Olivieri et al., 2020). From this data matrix, Webster was parameterized to infer 10 biological functions and approximate each gene effect as a sparse mixture of two functional effects.
(B) Top: the original fitness data, preprocessed to a set of 304 high-variance fitness gene effects from 31 treatment conditions, shown as a hierarchically clustered heatmap. Bottom: Webster’s approximation of the data, with each gene effect approximated as a sparse mixture of two inferred functions. The order of genes and treatments is preserved between panels.
(C) The dictionary matrix. Each column of the dictionary captures the inferred fitness effect of depleting an biological function learned from the data.
(D) The loadings matrix. Top: Sparse gene-to-function loadings for the 304 fitness genes. Each gene (column) has two nonzero loadings, encoding the model’s sparse representation of its gene effect. Bottom: Literature curated gene annotations, defined by Olivieri et al. (2020). Gene order is preserved between panels. TSG, tumor suppressor gene.
(E) Gene effect decomposition. Webster decomposes H2AFX knockout as a mixture of two functional effects related to DNA double-stranded breaks. The first function, homologous recombination, has a fitness effect induced by olaparib and camptothecin, etc. The second, end joining, has a fitness effect induced by doxorubicin and etoposide, etc. Webster faithfully modeled the H2AFX gene effect as the sum of these two functional effects, scaled by their respective loadings (Pearson = 0.76).
(F) Top: relationships learned from fitness data for H2AFX and MCPH1, an obligate H2AFX interactor. Each arrow corresponds to an inferred gene-to-function loading. Bottom: Illustration of H2AFX/MCPH1’s shared roles as DNA double-stranded break sensors upstream of homologous recombination and end joining.
(G) Additional gene loadings. Left: The top 15 genes ranked by their loadings on homologous recombination alongside literature annotations, as previously described in D. Right: Joint UMAP embedding of gene and functional effects, with genes colored by loading scores. Gene effect data from Olivieri et al. (2020). Functional effects inferred with Webster (this study) (see also Figure S2D).
(H) Same as (G), but for end joining gene loadings. In the UMAP, H2AFX and MCPH1 are embedded between homologous recombination and end joining. Gene effect data from Olivieri et al. (2020). Functional effects inferred with Webster (this study).
(I) A network of DNA damage functions, with the number of pleiotropic genes connecting functions represented by line thickness.