Figure 2: Exploiting phenotypic or genetic diversity in mouse populations to investigate molecular and genetic mediators of resilience.
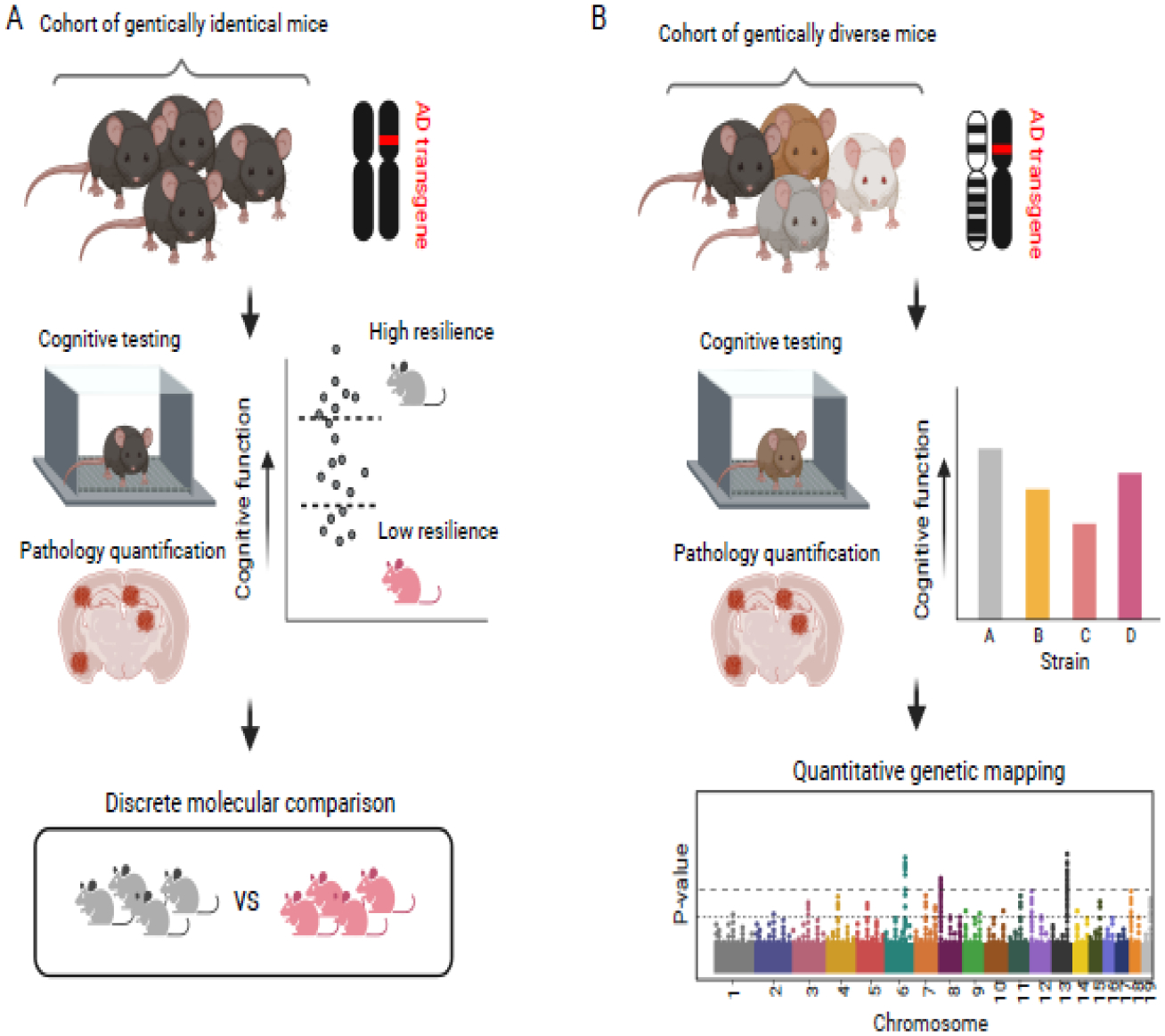
A) Phenotypic diversity among genetically homogenous strains of mice harboring Alzheimer’s disease (AD) transgenes can be harnessed to classify individuals into discrete ‘high resilience’ (i.e. strong-learner) or ‘low resilience’ (i.e. weak-learner) categories. Pathology can be quantified using a range of techniques including immunohistochemistry to account for differences in amyloid and/or hyperphosphorylated tau accumulation. B) Traditional animal models do not incorporate genetic diversity, which in human populations contributes to differing levels of resilience. By introducing AD transgenes into diverse models including the BXDs, Collaborative Cross, or Diversity Outbred panels, strain-by-strain differences in cognitive outcomes and pathology can be quantified and used for genetic mapping in order to identify specific genetic mediators of resilience.
