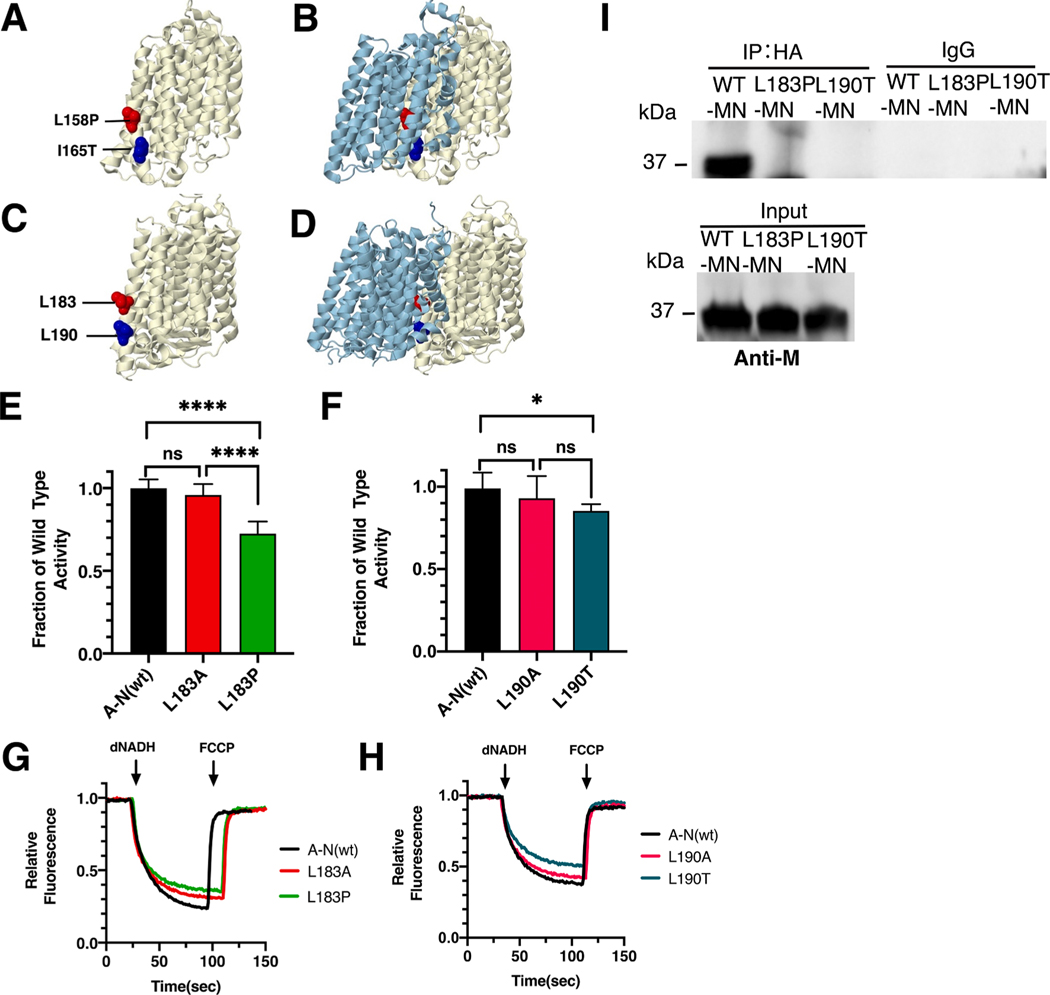
Fig. 4.
Analysis of ND4 mutations in nuoM of E. coli. (A) The locations of human mutations L158P (red) and I165T (blue) are shown in ND4 (light yellow), using PDB file 5xtd (Gu et al., 2016). (B) ND2 (light blue) is shown. (C) The locations of E. coli residues L183 (red) and L190 are shown in nuoM (light yellow), using PDB file 3rko (Efremov and Sazanov, 2011). (D) nuoN (light blue) is shown. (E, F) dNADH-oxidase activities of membrane vesicles prepared from the E. coli mutants are shown compared to a wild type sample prepared the same day. (G, H) Proton translocation rates from the same samples shown in panels E and F are indicated by fluorescence quenching of the acridine dye ACMA. (I) Membrane vesicles from E. coli cells carrying the modeled human mutations, M_L183P and M_L190T, in pBAD33-(MN), were prepared and solubilized with dodecyl maltoside. The samples, including pBAD33-(A-N), a control expressing the entire nuo operon, were immunoprecipitated with an HA antibody to bring down the HA-tagged subunit N. The input panel shows the presence of subunit M in all samples, while the IgG panel shows that in the absence of HA-antibody, no M subunits were precipitated. (E and F) ns, not significant; *P < 0.05; ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)