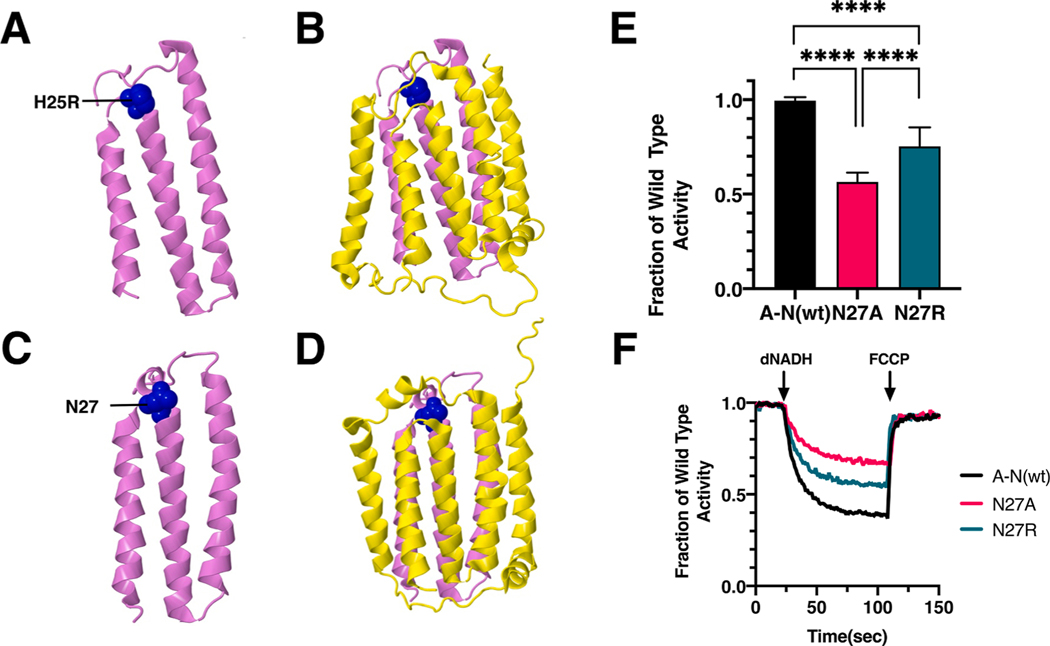
Fig. 8.
Analysis of an ND4L Mutation in nuoK of E. coli. (A) The location of human mutation H25R (blue) is shown in ND4L (violet), using PDB file 5xtd (Gu et al., 2016). (B) Neighboring subunit ND6 (yellow) is shown. (C) The location of E. coli residue N27 is shown in nuoK (violet), using PDB file 3rko (Efremov and Sazanov, 2011). (D) Neighboring subunit nuoJ (yellow) is shown. (E) dNADH-oxidase activities of membrane vesicles prepared from the E. coli mutants are shown compared to a wild type sample prepared the same day. (F) Proton translocation rates from the same samples shown in panel E are indicated by fluorescence quenching of the acridine dye ACMA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)