Figure 2. The PARP1-CycT1 binding depends on the histidine-rich domain of CycT1 and the PAR chains on PARP1 and is likely facilitated by electrostatic interactions between the two.
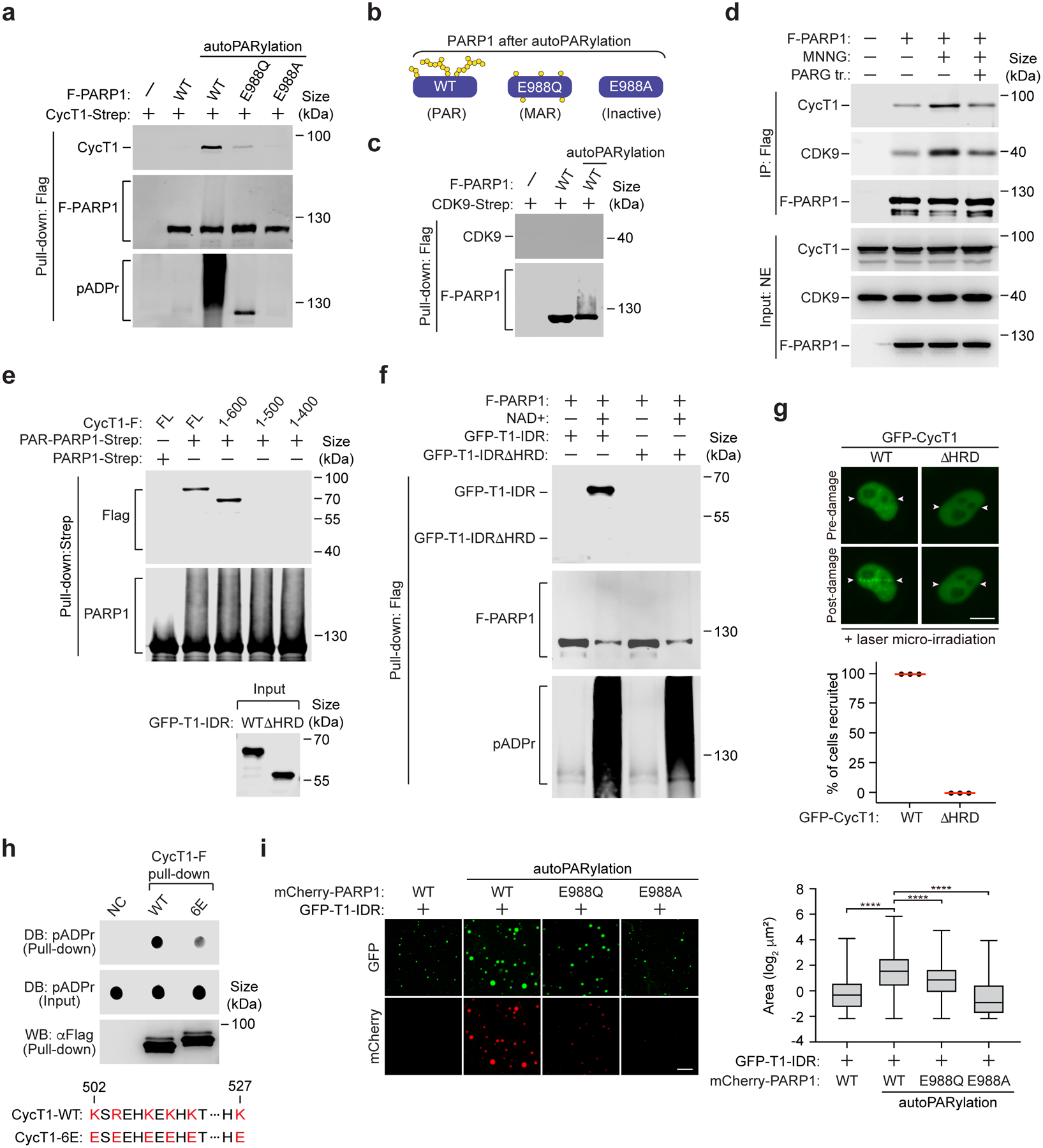
a, c, e, & f, Purified and immobilized WT or mutant F-PARP1 or PARP1-Strep was subjected to autoPARylation reaction in vitro. The pattern of ADP-ribose attached on WT and mutant PARP1 after the reaction is diagramed in b. After the reactions were stopped, the unmodified or modified PARP1 was incubated with purified CycT1-Strep (a), CDK9-Strep (c), full-length (FL) or truncated CycT1-F (e) or WT or the HRD-deleted GFP-CycT1-IDR (f). The input and pull-down proteins were analyzed by Western blotting (WB). d, HeLa cells expressing F-PARP1 or not were untreated or treated with MNNG. anti-Flag immunoprecipitates (IP) were incubated with recombinant PARG or not and then examined by WB. g, HeLa cells expressing WT or the HRD-deleted GFP-CycT1 were subjected to laser microirradiation and analyzed and quantified as in Fig.1i. Scale bar = 10 μM. The error bars indicate mean ± s.d. with n = 3 independent experiments. h, Purified and immobilized WT or mutant CycT1-F was incubated with autoPARylated PARP1. The input and pull-down proteins were analyzed by Dot blotting (DB) or WB. Bottom: The altered residues in CycT1–6E and their corresponding positions in WT CycT1 are in red. i, GFP-CycT1-IDR (0.4 mg/ml) mixed with unmodified mCherry-PARP1 or in vitro PARylated WT or mutant mCherry-PARP1 (0.1 mg/ml) was subjected to droplet formation assay and examined by fluorescence microscopy for GFP and mCherry fluorescence. Scale bar = 10 μM. Right: Size quantification of the droplets in each group. The box plots show the minimum, first quartile, median, third quartile and maximum with n representing the number of droplets examined over 3 independent experiments: mCherry-PARP1-WT (n=2907), mCherry-PARP1-WT-PARylation (n=2284), mCherry-PARP1-E988Q-PARylation (n=1797), mCherry-PARP1-E988A-PARylation (n=3782); statistical analysis was performed using two-tailed unpaired t-tests; ****P < 0.0001. All Western blots are representative of three independent experiments. Gel source data are available online.
