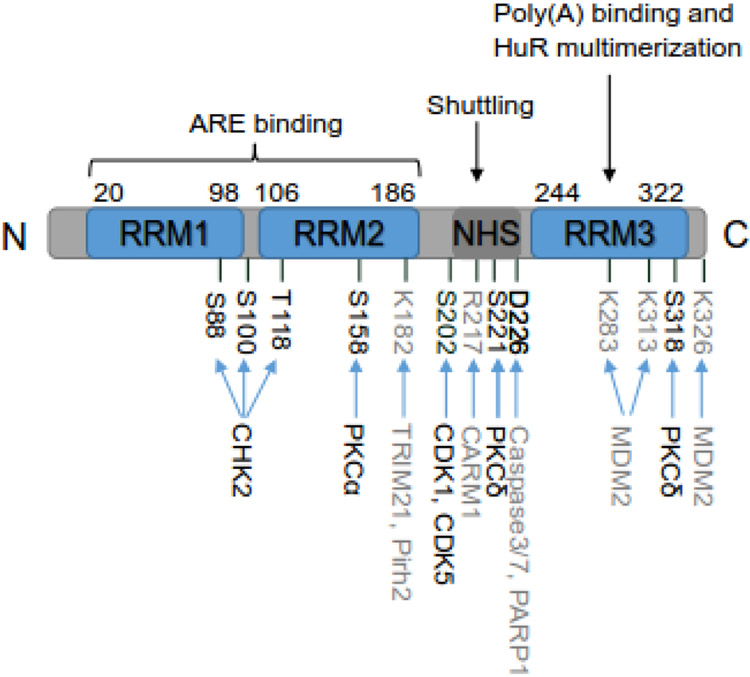
Figure 1:
Schematic structure of HuR. HuR protein consists of 326 amino acids (aa). It has three conserved RNA recognition motifs (RRMs) and an unconserved hinge region containing HuR nucleocytoplasmic shuttling (HNS) sequence. The amino acid positions of these regions are indicated. RRM1 and RRM2 are responsible for recognizing and binding to AREs of mRNA targets. RRM3 is involved in binding to poly(A) tail of mRNA targets and HuR multimerization on mRNA targets. The hinge domain mainly controls the nuclear and cytoplasmic shuttling of HuR. The major post-translational modified amino acids along with the corresponding enzymes are indicated as well, with phosphorylated amino acids and corresponding kinases in black and other modified amino acids and enzymes in grey.