Table 1.
Published small molecules that interfere with HuR–RNA interaction.
| ID | Structure | Binding affinity (IC50) |
Source | Ref |
|---|---|---|---|---|
| Quercetin |
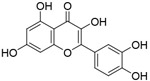
|
1.40μM | RNA EMSA screening of 179 chemicals | [93] |
| b-40 |
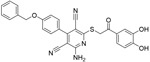
|
0.38μM | ||
| b-41 |
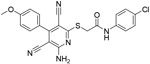
|
6.21μM | ||
| Mitoxantro ne |
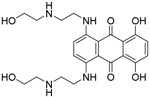
|
NA | AlphaScreen-based HTS of a chemical library of ~2000 small molecules | [95] |
| CMLD-1 |
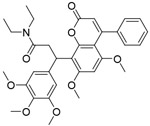
|
4.0μM | FP-based HTS of ~6000 compounds from CMLD library and a library of FDA-approved drugs | [94] |
| CMLD-2 |
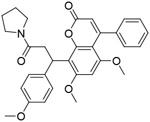
|
2.4μM | ||
| Dihydrotanshinone-l |
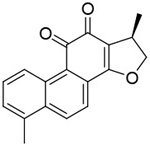
|
68nM | AlphaScreen-based HTS of 107 anti-nflammatory compounds | [101] |
| C10 |
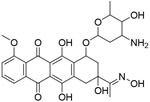
|
NA | FP-based HTS of 1597 compounds from NCI diversity set V library | [125] |
| AZA-9 |
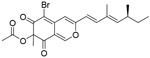
|
1.2μM | FP-based HTS of ~2000 compounds from the NCI library and inhouse compounds | [126] |
| 6a |
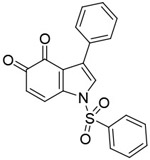
|
12.8nM (Ki) | rationally designed new chemicals | [102] |
| 6n |
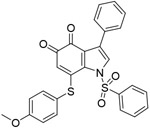
|
15nM (Ki) | ||
| Compoun d 4 |
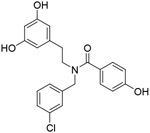
|
NA | rationally designed new chemical | [127] |
| KH-3 |
|
3.54μM | FP-based HTS of ~2000 compounds from the NCI library and inhouse compounds | [50] |
| VP12/14 |
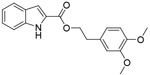
|
NA | rationally designed new chemicals | [128] |
| VP12/110 |
|
NA | ||
| Compound 2 |
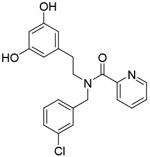
|
105μM | rationally designed new chemicals | [129] |
| Compound 3 |
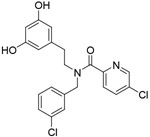
|
92μM |
NA: not available
