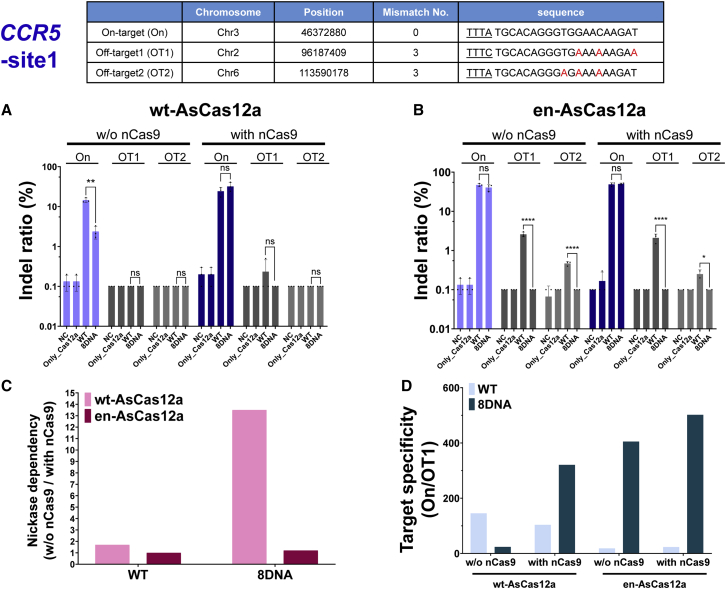
Figure 3.
Comparison of genome-editing specificity of en-AsCas12a and wt-AsCas12a based on chimeric DNA-RNA guide targeting endogenous locus in cell lines (HEK293FT)
Upper table shows the on-target nucleotide sequence (On) for target gene (CCR5-site1) and the corresponding off-target nucleotide sequence (OT1, OT2) predicted from in silico analysis (Bae et al. 31). Underlining indicates the PAM (TTTN) nucleotide sequence, and the nucleotides mismatched with the target sequence in the off-target are indicated in red. (A and B) Indel ratio (%) of the wt-AsCas12a-based (A) or en-AsCas12a-based (B) editing on the endogenous target sequences (on-/off-target sites for CCR5-site1) using wt-crRNA (WT) and 3′ end 8 nt DNA-substituted crRNA (8DNA). NC, negative control; Only_Cas12a, only protein treated; nCas9, nickase Cas9 (D10A). (C and D) Nickase dependency (C) and target specificity (D) were calculated from next-generation sequencing results. Nickase dependency = (without [w/o] nCas9 editing [%]/with [w/] nCas9 editing [%]); Target specificity = (on-target editing [%]/off-target editing [%]). Histograms represent means ± SEM from three independent experimental values. p values were calculated using two-way ANOVA with Sidak’s multiple comparisons test (ns, not significant; ∗p < 0.0332, ∗∗p < 0.0021, ∗∗∗p < 0.0002, ∗∗∗∗p < 0.0001).