Figure 2.
FBW2 assembles an SCF complex and interacts with AGO1 in planta
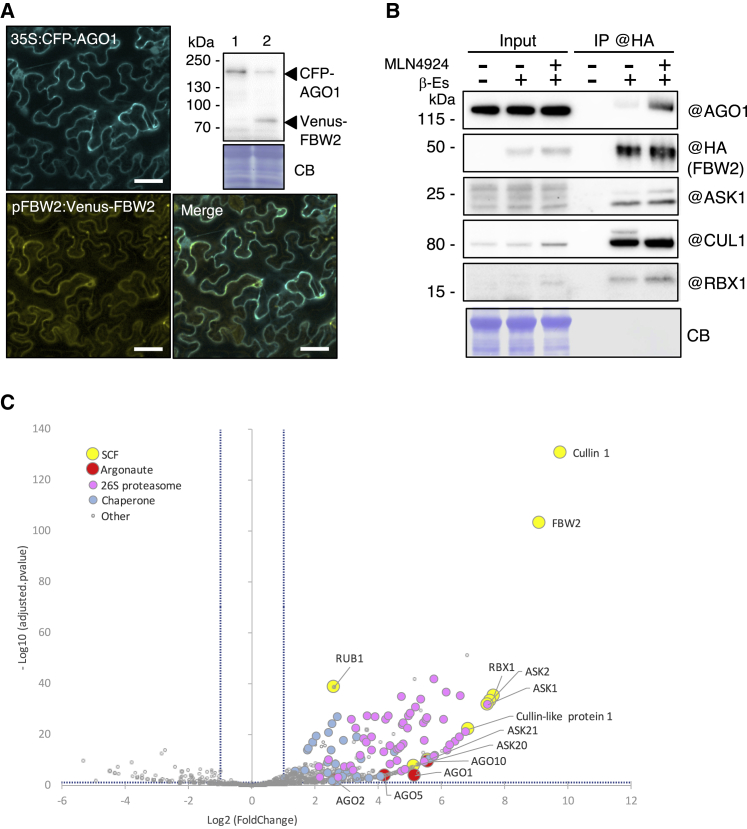
(A) Subcellular localization of CFP-AGO1 and Venus-FBW2 by confocal microscopy. Co-infiltration of 4-week-old N. benthamiana leaves with agrobacteria-harboring binary vectors for the expression of fluorescent-tagged protein constructs. Bacteria were infiltrated at an OD of 0.1. Pictures were taken and sampled 3 days later. For confocal microscopy imaging, CFP and Venus were excited at 458 and 514 nm, respectively. Emission signals were recovered between 465 and 510 nm for the CFP and 520 and 596 nm for the Venus. Scale bars, 40 μm. Immunodetection using GFP antibodies of protein extracts from agro-infiltrated leaves with 35S:CFP-AGO1 and pFBW2:Venus-FBW2 (lane 2) constructs is included. Expression of GUS (lane 1) served as negative control. CB staining was used as a loading control.
(B) FBW2 assembles an SCF complex and interacts in planta with AGO1 (based on two biological replicates; see also Figure S9). Western blot of protein extracts from 10-day-old XVE:3HA-FBW2 seedlings. 3HA-FBW2 was immunoprecipitated with anti-HA antibodies after an overnight induction of expression in liquid MS medium supplemented with DMSO (−) or β-Es (10 μM, +). 3HA-FBW2 co-immunoprecipitates with SCF components, ASK1, CUL1, and RBX1. Blocking the SCF activity with the drug MLN4924 further allowed co-immunoprecipitation of AGO1. @ indicates hybridization with the corresponding antibodies.
(C) FBW2 interactome revealed by immunoprecipitation and mass spectrometry. We compared eight samples (4 samples of FBW2OE and 4 samples of FBW2OE/ago1-27) from two independent biological replicates to seven control samples. Volcano plot shows the enrichment of proteins co-purified with HA-tagged FBW2 bait compared with Col-0 controls. The y and x axes display log values from adjusted p values and fold changes, respectively. The horizontal dashed line indicates the threshold above which proteins are significantly enriched (adjusted p values < 0.05). The vertical dashed lines indicate the fold change thresholds for FBW2-enriched proteins (log2 > 1) or Col-0-enriched proteins (log2 < −1). Four color-coded functional clusters are highlighted in the case of proteins enriched in the FBW2 coIP samples. The source data are available in Table S1.