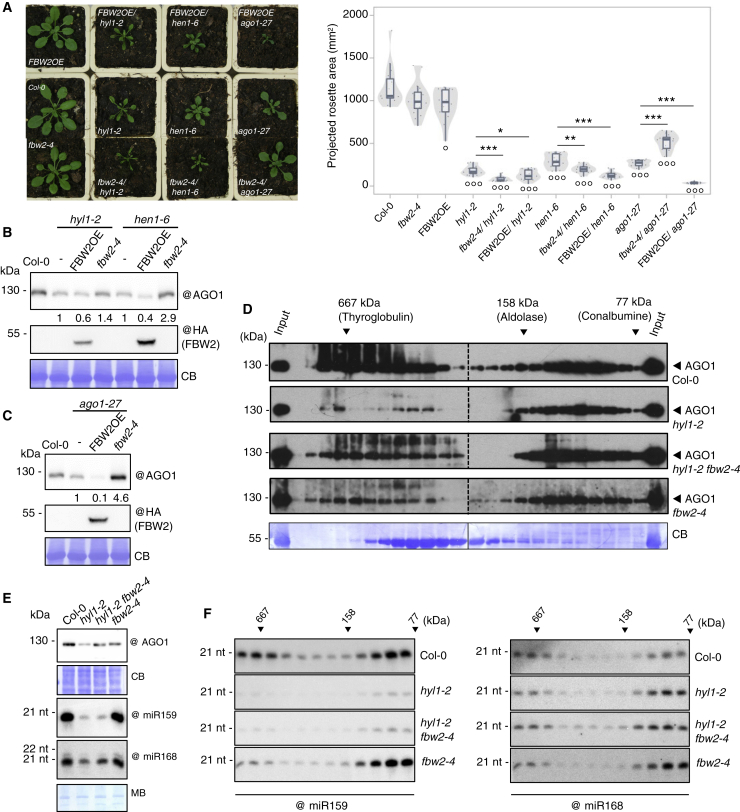
Figure 4.
Effects of FBW2 overexpression or loss of function in silencing mutants restores high-molecular-weight AGO1 complexes
(A) Representative pictures of 27-day-old plants of Col-0, hyl1-2, hen1-6, ago1-27, and their crosses with fbw2-4 or 35S:3HA-FBW2 (FBW2OE) as indicated (based on two biological replicates; see also Figure S3A). Right: quantitative analysis of the experiment represented on the left, with n > 12 plants per genotype. °p < 0.05 and °°°p < 0.001 (Student’s t test) compared with Col-0. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Student’s t test) compared with the corresponding single mutant.
(B and C) Western blot of protein extracts from the same seedlings as indicated in Figure S3A (for biological replicates see Figures 5 and S3–S5). CB staining was used as loading control. AGO1 signal was quantified by ImageJ, normalized to the corresponding CB. Numbers below the panel indicate relative to the corresponding mutants (hyl1-2, hen1-6 and ago1-27, respectively) set at 1.0. @ indicates hybridization with the corresponding antibodies.
(D and E) Gel filtration analysis of AGO1-based RISC complexes in Col-0, hyl1-2, hy1-2 fbw2-4, and fbw2-4 13-day-old seedlings (a biological replicate is shown in Figure S4). Proteins of known molecular weight are shown on top of the blot. CB staining was used as loading control and “@” indicates hybridization with the AGO1 antibody. (E) Shown are protein and sRNA analysis of the input fraction prior to gel filtration. Methylene blue (MB) staining of the membrane was used as loading control.
(F) sRNA analysis from even fractions, spanning the same range (from the GF of [D]). For this analysis, 10μg of the RNA per lane was loaded. The “@” symbol indicates hybridization with the indicated oligonucleotide probes.