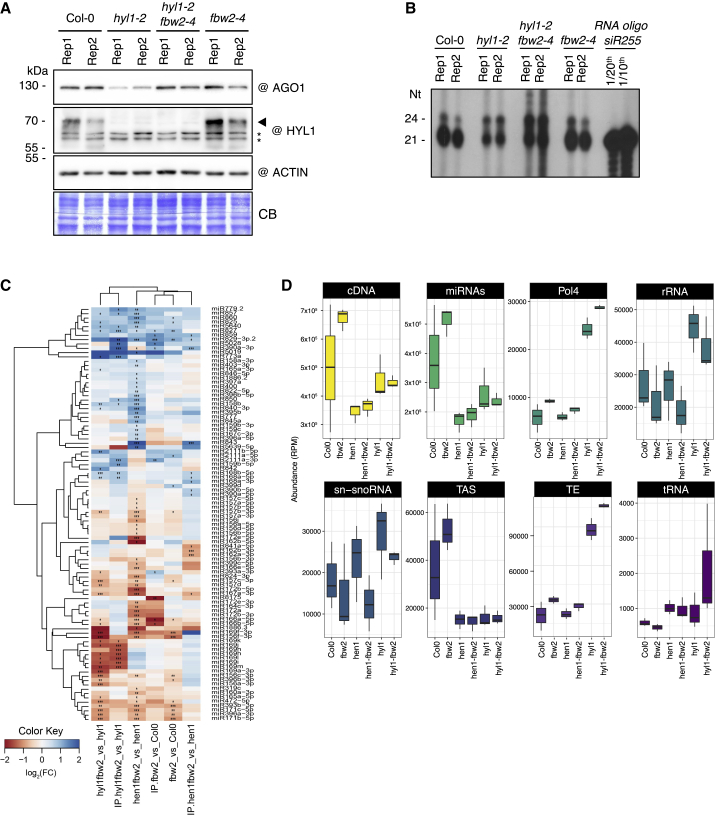
Figure 5.
Loss of FBW2 modifies AGO1 loading in hyl1-2
(A) Western blot of total protein extracts from 2-week-old seedlings from Col-0, hyl1-2, hy1-2 fbw2-4, and fbw2-4 mutants. Two biological replicates (1 and 2) are shown. CB staining and ACTIN were used as loading controls, and @ indicates hybridization with the corresponding antibodies. The arrow indicates the HYL1 protein band, and ∗ indicates aspecific cross-reacting bands.
(B) Denaturing polyacrylamide gel of sRNA from immunoprecipitated AGO1 (based on two biological replicates). RNAs from the same protein extracts shown in (A) were indiscriminately labeled by replacing their 5′ phosphate with a radioactive one using polynucleotide kinase (PNK). An oligo corresponding to the siR255 serves as control for sRNA size.
(C and D) Deep-sequencing analyses of total and AGO1-IP sRNA (performed on three biological replicates). (C) Relative abundance of miRNA in AGO1 IP samples with significant differential expression in single and double mutants compared with WT (Col-0) and single mutants; the relative abundance is expressed as a heatmap (see Key at the bottom), with the samples being compared indicated below each heatmap (∗Q value #0.05, ∗∗Q value #0.01, and ∗∗Q value #0.001). (D) Boxplot representing the abundance of reads (in reads per million, RPM) mapping to eight different features of the Arabidopsis genome TAIR 10 in AGO1 IP samples. These include the following, from left to right: cDNA; mature miRNA; siRNA precursors dependent on Pol4; ribosomal RNAs (rRNAs); small nuclear and small nucleolar RNA (snRNA and snoRNA); TAS precursors; transposable elements (TEs); and tRNA-derived sRNA (tRNA).