Figure 6.
Loss of FBW2 modifies sRNA-mediated targeting in hyl1-2
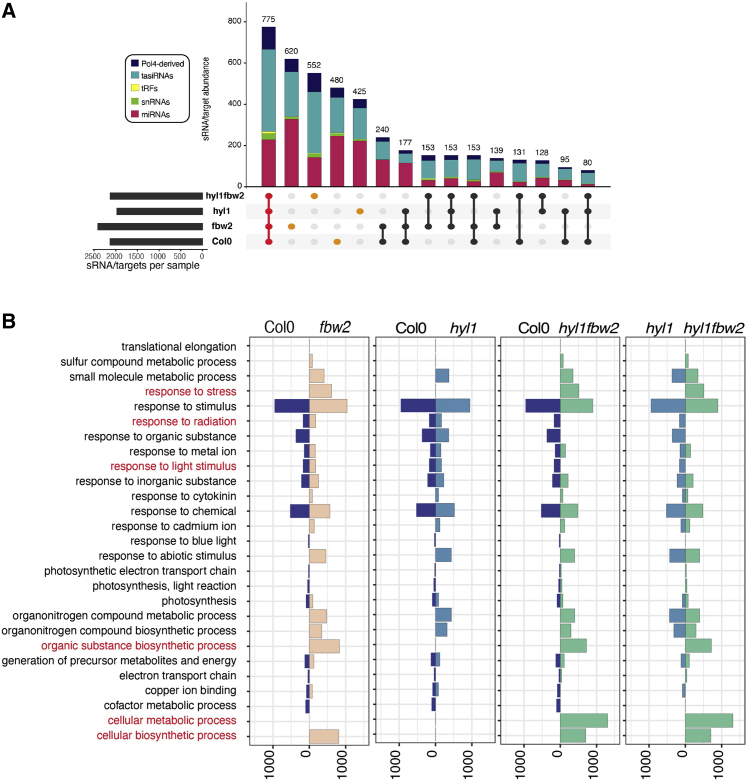
(A) Deep-sequencing analyses of PARE (performed on the same biological replicates as in Figures 5C and 5D). Visualization of the intersection of sRNA/target signatures among the different mutants, represented with an UpSet plot. Top: a vertical bar plot with the number of signatures included in each intersection, color coded into five sRNA categories, including, from top to bottom, siRNA precursors dependent on Pol4; siRNA from TAS precursors (tasiRNA); tRNA-derived sRNA (tRF); small nuclear and small nucleolar RNA (snRNA); and miRNA. Bottom left:a horizontal bar chart with the number of signatures included in each set. Bottom right indicates which intersections and their aggregates are being considered in each case. Labeled in red are the signatures common to all samples, and in orange are the signatures unique to each mutant.
(B) Vertical bar plot representing the number of genes (horizontal scale) included in each gene ontology (GO) term (vertical scale). Only significantly enriched GO terms are represented for each of the tested genotypes. From left to right are shown Col-0 versus fbw2 mutants, Col-0 versus hyl1 mutants, Col-0 versus hyl1 fw2 double mutants, and hyl1 mutants versus hyl1 fbw2 double mutants.