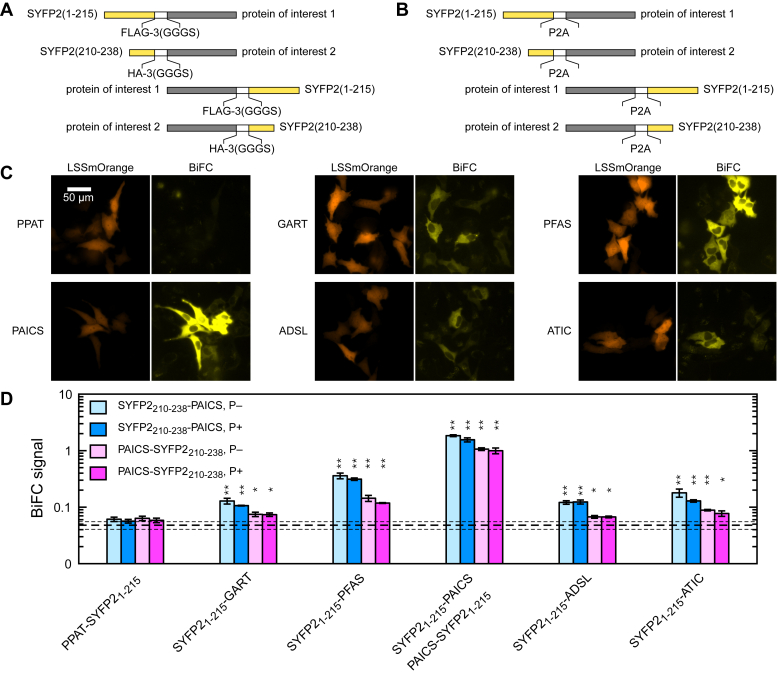
Figure 2.
Probing PPIs between PAICS and other DNPB enzymes using BiFC. Design schematic for plasmid expression cassettes encoding (A) BiFC assay and (B) BiFC control fusion constructs. For the BiFC control constructs, the linker between the SYFP2 fragment and the DNPB enzyme is replaced with a self-cleaving P2A peptide. C, representative images showing the reconstituted SYFP2 signal in HeLa cells cultured due to the interaction of SYFP2210-238-PAICS with PPAT-SYFP21-215, SYFP21-215-GART, SYFP21-215-PFAS, SYFP21-215-PAICS, SYFP21-215-ADSL, and SYFP21-215-ATIC. Here, LSSmOrange labels cells that have been successfully transfected with the BiFC expression vectors. D, quantitated BiFC signal in HeLa cells between PAICS with all the DNPB enzymes. Dashed lines indicate background BiFC signal ± standard deviation, as obtained from self-cleaving P2A control constructs. p-values from unpaired t tests comparing BiFC signal versus P2A controls: ∗ <0.01, ∗∗ <0.001. ADSL, adenylosuccinate lyase; ATIC, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase; BiFC, bimolecular fluorescence complementation; DNPB, de novo purine biosynthesis; GART, phosphoribosylglycinamide synthetase/formyltransferase/phosphoribosylaminoimidazole synthetase; PAICS, phosphoribosylaminoimidazole carboxylase/succinocarboxamide synthetase; PFAS, phosphoribosylformylglycinamidine synthase; PPI, protein–protein interaction; PPAT, amidophosphoribosyltransferase.