Figure 1.
Patient-derived DM1 and DM2 fibroblast lines display molecular markers of disease.
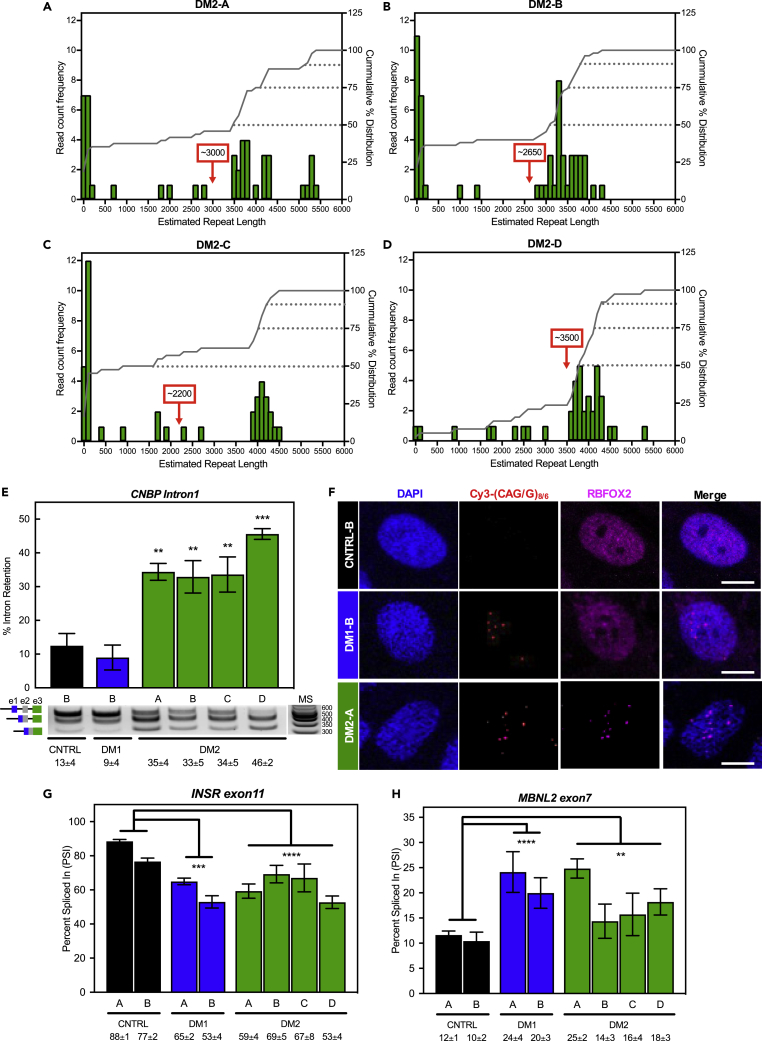
Observed Bionano reads (A–D) are shown for all DM2 fibroblast lines. Histograms of the estimated CCTG repeat lengths (green bars) are shown as read count frequency (left y axis) with the cumulative % distribution functions on the right (gray line, right y axis). The 50th, 75th, and 90th percentiles of repeat lengths are indicated with gray dotted lines. The average repeat size is displayed in the red box with arrow. Note the lack of a large peak at the shorter unaffected repeat size for the homozygous DM2-D cell line (D).
(E) Intron retention assay (E) demonstrated significant CNBP intron 1 retention in all DM2 (green) cell lines compared to control cell lines (black), but not in DM1 cells (blue). Representative gel image of PCR products using primer set amplifying intron 1 to exon3 of CNBP (product schematic shown left of gel image and molecular standard (MS) values are in base pairs to the right). Mean % intron retention ±SD is denoted below (two-tailed t-test, ∗∗p < 0.01, ∗∗∗p < 0.001).
(F) Fluorescence in situ hybridization immunofluorescence microscopy (F) in control, DM1, and DM2 fibroblasts against (C)CUG RNA using either Cy3-(CAG)8 or -(CAGG)6 probes (red). RBFOX2 immunofluorescence shown in purple and DAPI shown in blue. All images were taken using the same laser and contrast/processing settings and a representative scale bar (10μm) is included with each set of micrographs.
(G and H) Splicing analysis (G and H) of control, DM1 and DM2 fibroblasts for INSR exon 11 (G) and MBNL2 exon7 (H) alternative exon events expressed as percent spliced in (PSI). Mean % spliced in ±SD is denoted below (ANOVA two-tailed test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). All experiments were done in at least triplicate using biological replicates.