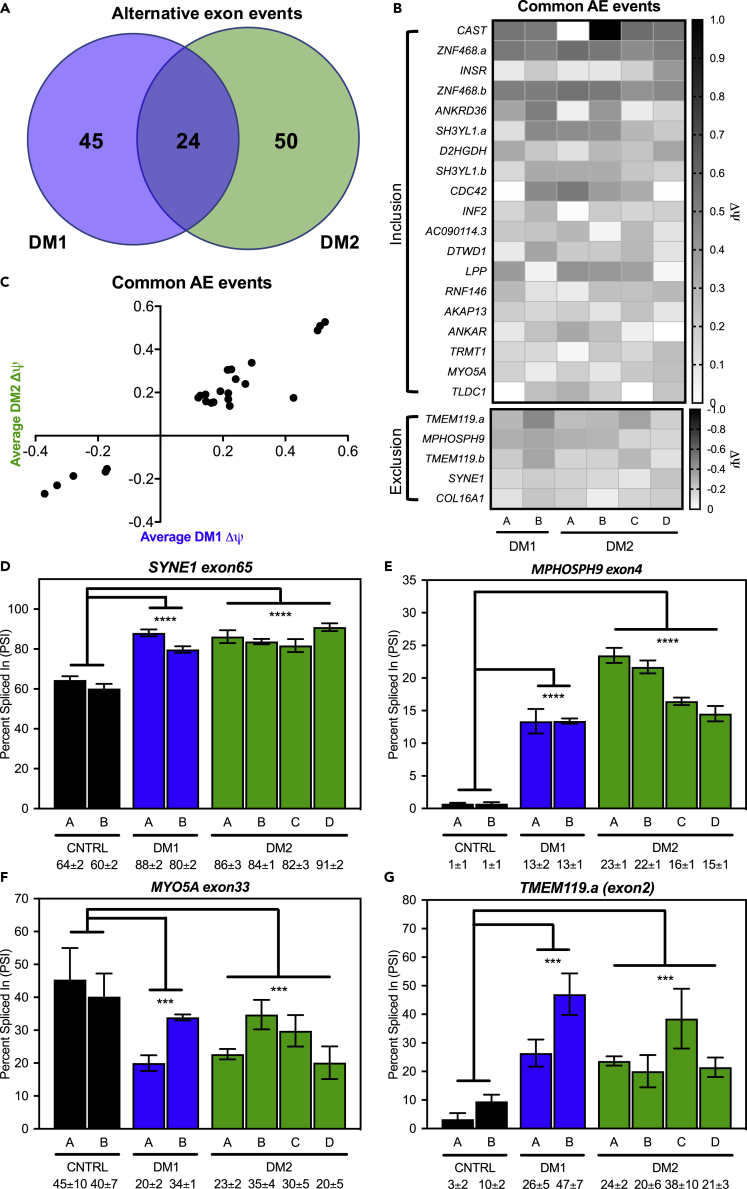
Figure 2.
Global mis-splicing occurs in DM1 and DM2 fibroblasts
Splicing analysis of alternative exon events determined via RNA-seq identified both unique and overlapping events in DM1 (blue) and DM2 (green) fibroblasts.
(A) (FDR<0.1, ΔΨ ≥ 0.1, ≥25 reads across exon junction).
(B) Heatmap of change in percent spliced-in (ΔΨ) for 24 alternative exon events shared between DM1 and DM2 fibroblasts (B) shows bias toward inclusion (19 events) over exclusion (5 events).
(C) Plot of average DM1 ΔΨ (x axis) versus average DM2 ΔΨ for the 22 common alternative exon events (C) shows similar extents of mis-splicing between DM1 and DM2.
(D–G) RT-PCR splicing analysis of control, DM1, and DM2 fibroblasts for common alternative exon events in SYNE1 exon65 (D), MPHOSPH9 exon4 (E), MYO5A exon33 (F), and TMEM119.a (exon2) (G) expressed as percent spliced in (PSI). Mean % spliced in ±SD is denoted below (ANOVA two-tailed test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). All experiments were done in at least triplicate using biological replicates.