Figure 4.
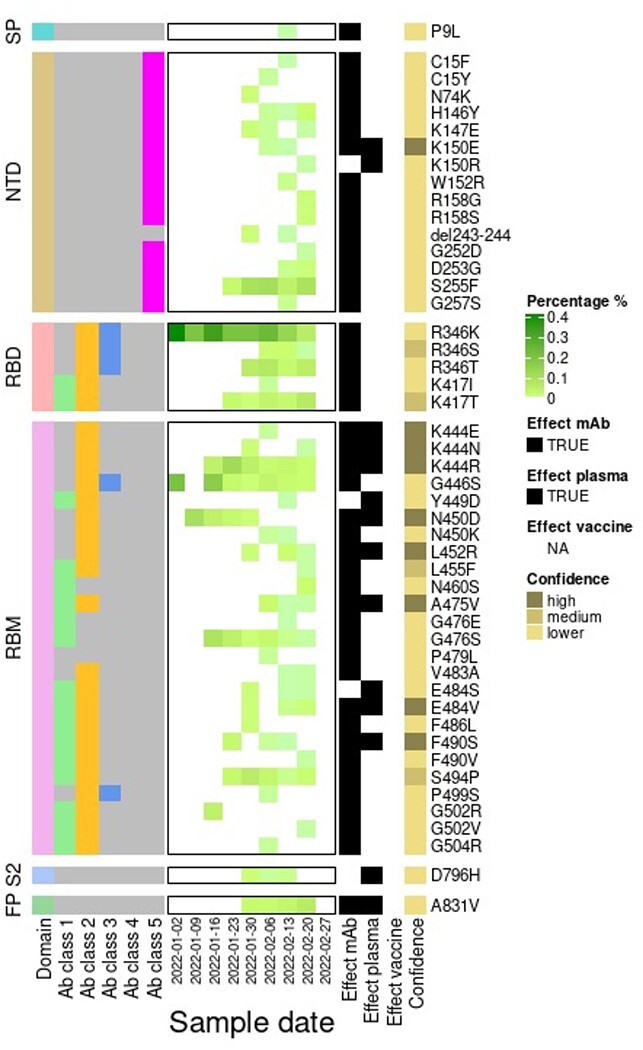
Heatmap showing the frequency of spike amino acid substitutions and a deletion with a potential or confirmed antigenic role on top of BA.2 through time. The labelled structural domains are indicated on the left side: SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor-binding motif; S2, subunit; FP, fusion peptide. Residues are also coloured according to the class of antibody that binds to an epitope. RBD antibody Classes 1–4 (Barnes et al. 2020) are depicted by colours: green (Class 1: ACE2 blocking, bind open RBD only), yellow (Class 2: ACE2 blocking, bind open, and closed RBD), blue (Class 3: non-ACE2 blocking, bind open, and closed RBD), or yellow (Class 4: non-ACE2 blocking, bind open RBD only). Residues described in an NTD epitope (Chi et al. 2020) are coloured in magenta (Class 5). Each residue is also classified as having evidence for mutations either affecting neutralisation by mAbs (Baum et al. 2020; Li et al. 2020; Weisblum et al. 2020; Liu et al. 2021) or serum from previously infected individuals (convalescent plasma) (Li et al. 2020; Weisblum et al. 2020; Andreano et al. 2021; Greaney et al. 2021; Liu et al. 2021) or vaccinated individuals (Wang et al. 2021) and emerging upon exposure to mAbs (Baum et al. 2020; Weisblum et al. 2020; Liu et al. 2021) or plasma (Weisblum et al. 2020; Andreano et al. 2021) in laboratory experiments.
