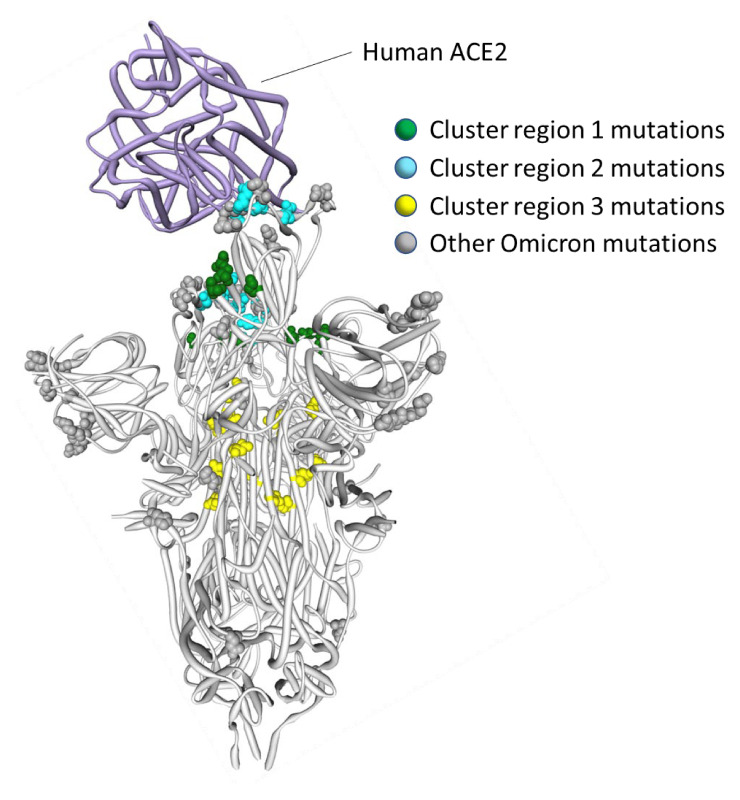
Fig. 5.
Positions on the three-dimensional SARS-CoV-2 Spike trimer of amino acids encoded by three clusters of BA.1 codon sites that are evolving either neutrally or under negative selection in non-Omicron SARS-CoV-2 sequences. The Spike protomer subunit interacting with human ACE2 is in the “up” configuration and the other two are in the “down” configuration (Xu et al. 2021). The cluster region 1 and 2 encoded amino acid changes in BA.1 (in green and blue, respectively) are within the receptor-binding domain of Spike with the cluster 2 encoded changes located within the receptor-binding motif. The cluster region 3 mutations are within the fusion domain of Spike. An interactive version of this figure can be found at https://observablehq.com/@stephenshank/sc2-omicron-clusters.