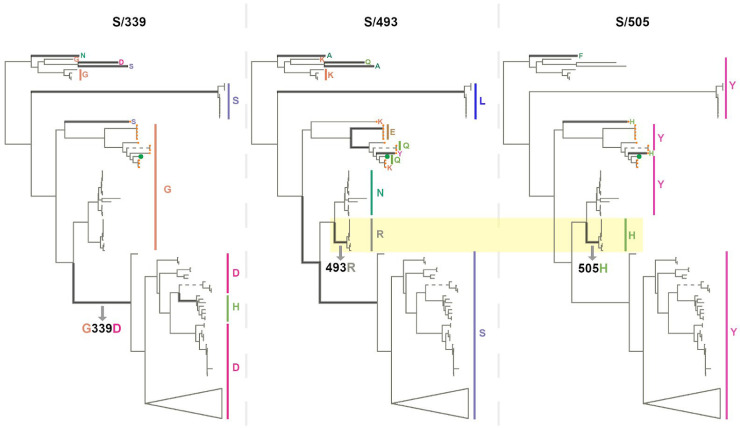
Fig. 6.
Phylogenetic trees of 167 sarbecoviruses indicating patterns of selection at S-gene codons S/339 (left tree), S/493 (middle tree), and S/505 (right tree). Branches along which amino acid states have changed are indicated with thick lines. Dashed lines represent long branches that have been shortened for visual clarity. The highlighted segments of the middle and right trees indicate the branch across which S/N493R and S/Y505H mutations occurred. The trees represent evolutionary relationships between putatively nonrecombinant sequence fragments in the genome region corresponding to Wuhan-Hu-1 Spike positions 324-654. The clade containing sarbecoviruses sampled in Europe and Africa has been used as the outgroup for rooting. Tree tips are annotated by amino acid states at the respective sites. SARS-CoV-2 is annotated with a green tip symbol and the nCoV clade sequences with a tip symbol in orange.