Figure 2.
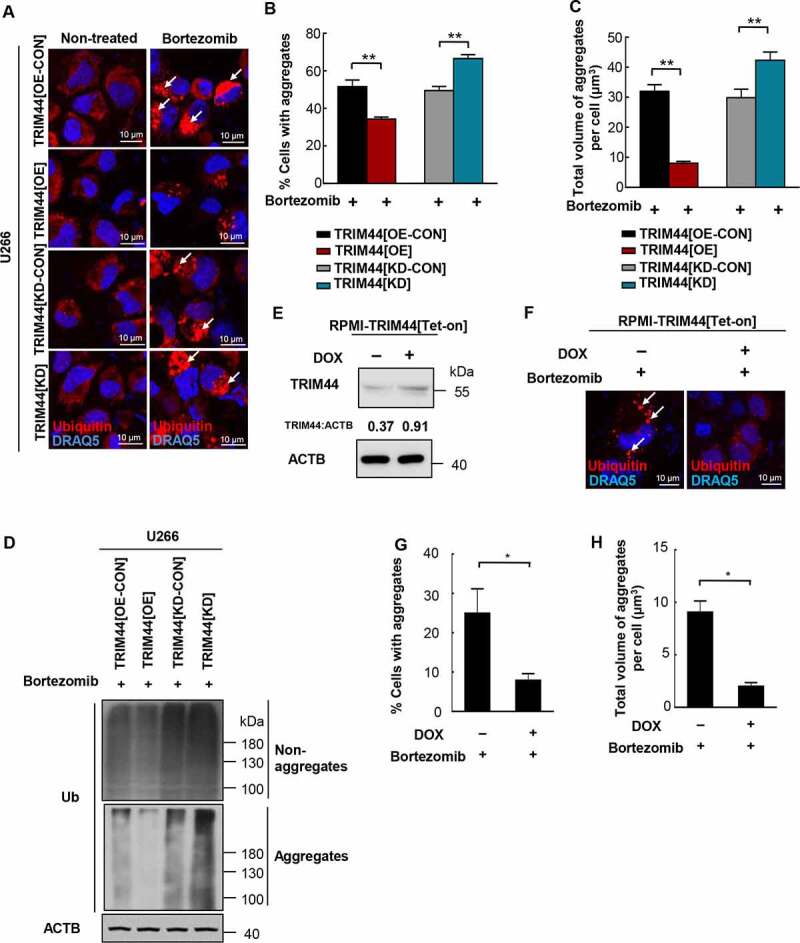
TRIM44 is required for aggregates deaggregation and clearance in MM cells. (a, b) U266 cells (TRIM44[OE-CON], TRIM44[OE], TRIM44[KD-CON] and TRIM44[KD]) were treated withor without bortezomib (5 nM) for 24 h to induce aggregates formation. Aggregates (marked by arrows) were identified by staining with the antibody against ubiquitin (a). Scale bars: 10 μm. The status of aggregates after MG132 treatment was quantified in the histogram (b). *, P < 0.05; **, P < 0.01. (c) Quantification of the total volume of aggregates in U266 cells (TRIM44[OE-CON], TRIM44[OE], TRIM44[KD-CON] and TRIM44[KD]). The volume of aggregates in 100 cells was counted for each data point shown. (d) Western blot analysis of non-aggregates and aggregates fractions of U266 cells (TRIM44[OE-CON], TRIM44[OE], TRIM44[KD-CON] and TRIM44[KD]) treated with bortezomib (5 nM, 24 h) and probe with indicated antibodies. Non-aggregates and aggregates protein were fractionated using NP40 lysis buffer. After centrifugation, the supernatant was used as non-aggregates fraction and the pellet was extracted in 1% SDS to obtain aggregates protein fraction. (e-g) RPMI-TRIM44[Tet-on] cells were treated with or without DOX (1 µg/mL, 24 h) together with bortezomib (10 nM, 24 h) treatment to induce aggregates formation. Aggregates (marked by arrows) were identified by staining with the antibody against ubiquitin. Scale bars: 10 μm. The protein level of TRIM44 were assayed by western blot (e). The status of aggregates after bortezomib treatment was quantified in the histogram (g). *, P < 0.05. (h) Quantification of the total volume of aggregates in RPMI-TRIM44[Tet-on] cells treated with or without DOX (1 µg/mL, 24 h) together with bortezomib (10 nM, 24 h) treatment to induce aggregates formation. The volume of aggregates in 100 cells was counted for each data point shown. *, P < 0.05.
