Extended Data Fig. 5. Differentially accessible promoters across pseudo-states.
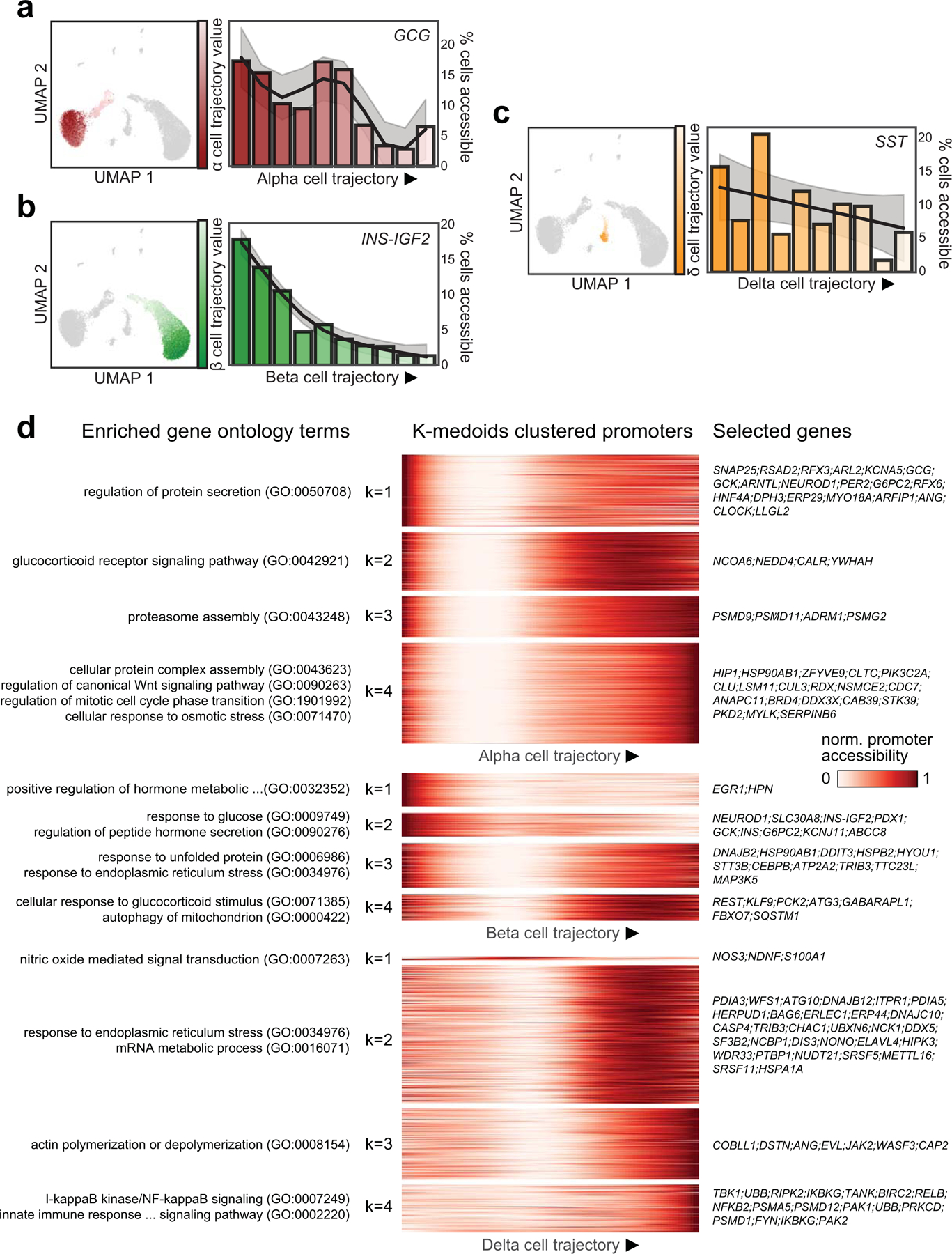
(a) Pseudo-state (trajectory) values for alpha cells plotted on UMAP coordinates (left) and percentage of cells with GCG promoter accessibility decreases across 10 bins along the alpha (α) cell trajectory (right). (b) Pseudo-state (trajectory) values for beta (β) cells plotted on UMAP coordinates (left) and percentage of cells with INS promoter accessibility decreases across 10 bins along the beta cell trajectory (right). (c) Pseudo-state (trajectory) values for delta (δ) cells plotted on UMAP coordinates (left) and percentage of cells with SST promoter accessibility decreases across 10 bins along the beta cell trajectory (right). (d) Heatmaps showing promoters with dynamic accessibility across trajectories for alpha (top), beta (middle) and delta (bottom) cell trajectories. Gene promoters are clustered into 4 groups for each trajectory with k-medoids clustering. Enriched gene ontology for each k-medoid cluster (left) and selected genes present in at least one enriched gene ontology.
