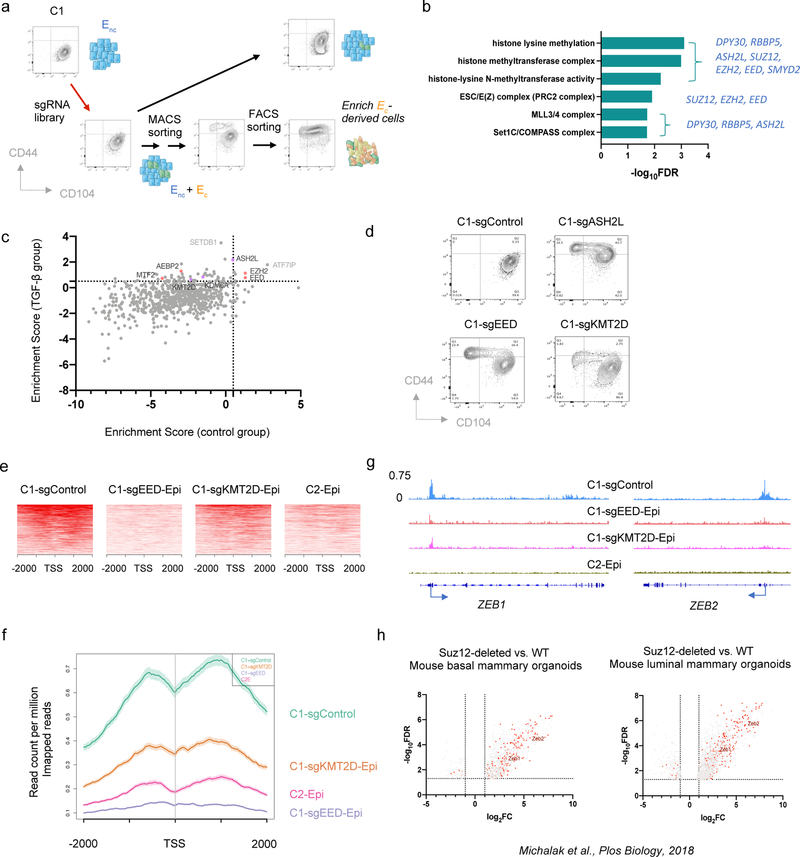
Figure 2. CRISPR screening identifies PRC2 and KMT2D-COMPASS as regulators of EMP.
a, Diagram of the CRISPR screening using non-convertible C1 cells to identify potential regulators of EMP. Enc, non-convertible epithelial cells. Ec, convertible epithelial cells. b, List of GO terms that were enriched in identified genes from the genome-wide CRISPR screening as guardians of the stable epithelial state. c, Plot showing the enrichment scores of genes examined using the EPIKOL CRISPR screening. Red and Purple dots indicate PRC2 and KMT2D-COMPASS components respectively. d, Flow cytometry analysis of the CD44 and CD104 cell-surface staining of single cell clones of C1-derived cells with control guide RNA or complete knock-out of ASH2L, EED or KMT2D genes. e, Heatmap displaying PRC2 occupancy (as measured by EZH2 CUT&RUN profiles) at gene promoters in C1-sgControl, C1-sgEED-Epi, C1-sgKMT2D-Epi and C2-Epi cells. 998 identified PRC2 direct target genes were shown in the plots. f, Average binding intensity of PRC2 in the promoter region of identified targets in C1-sgControl, C1-sgEED-Epi, C1-sgKMT2D-Epi and C2-Epi cells. The error bands represent the standard error of mean. g, Status of PRC2 occupancy at the promoters of EMT-TF genes ZEB1 and ZEB2, signal quantified as counts per million mapped reads. h, ZEB1 and ZEB2 were up-regulated in mouse epithelial cells after PRC2 core component SUZ12 knock-out. Red dots represent genes identified as PRC2 direct targets in HMLER-C1 cells. Numerical source data are provided.