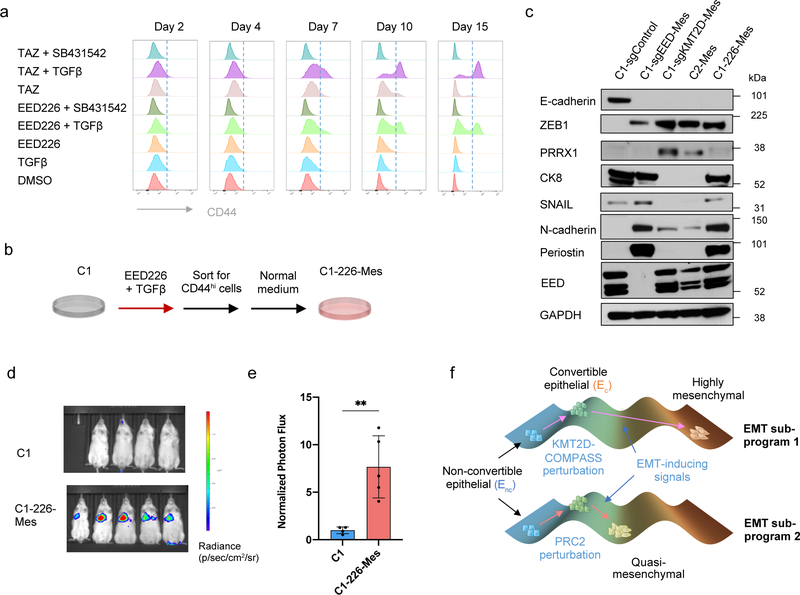
Figure 6. Transient inhibition of PRC2 is sufficient to generate a metastatic, quasi-mesenchymal cell state.
a, Time-course flow cytometry analysis of the CD44 cell-surface staining of C1 cells treated with different combinations of TGF-β (2ng/ml), SB-431542 (5μM), EED226 (10μM) and Tazemetostat (TAZ) (10μM). b, C1–226-Mes cells were generated by treating C1 cells with EED226 and TGF-β for 10 days and then FACS-sorting the CD44hi population. c, Immunoblot of PRC2 component EED, EMT-TFs SNAIL, ZEB1, PRRX1 and EMT markers E-cadherin, Keratin 8, N-cadherin and Periostin in C1-sgControl, C1-sgEED-Mes, C1-sgKMT2D-Mes cells, C2-Mes and C1–226-Mes cells. n = 2 biologically independent experiments. d,e, Mice images (d) and quantification of bioluminescence (e) of mice intravenously injected with parental C1 or C1–226-Mes cells expressing luciferase reporter. Data were collected 14 days after cell injection. n=5. **, p = 0.005. Statistical analysis was performed using unpaired two-tailed Student t-tests. Data are presented as mean ± SEM. f, Schematic representation of the model in which loss of PRC2 and KMT2D-COMPASS enables EMP and specifies two EMT subprograms to generates distinct mesenchymal cell states. Numerical source data are provided.