Abstract
Plants possess regulatory mechanisms that allow them to flower under conditions that maximize reproductive success. Selection of natural variants affecting those mechanisms has been critical in agriculture to modulate the flowering response of crops to specific environments and to increase yield. In the temperate cereals, wheat and barley, the photoperiod and vernalization pathways explain most of the natural variation in flowering time. However, other pathways also participate in fine-tuning the flowering response. In this work, we integrate the conserved microRNA miR172 and its targets APETALA2-like (AP2L) genes into the temperate grass flowering network involving VERNALIZATION 1 (VRN1), VRN2 and FLOWERING LOCUS T 1 (FT1 = VRN3) genes. Using mutants, transgenics and different growing conditions, we show that miR172 promotes flowering in wheat, while its target genes AP2L1 (TaTOE1) and AP2L5 (Q) act as flowering repressors. Moreover, we reveal that the miR172-AP2L pathway regulates FT1 expression in the leaves, and that this regulation is independent of VRN2 and VRN1. In addition, we show that the miR172-AP2L module and flowering are both controlled by plant age through miR156 in spring cultivars. However, in winter cultivars, flowering and the regulation of AP2L1 expression are decoupled from miR156 downregulation with age, and induction of VRN1 by vernalization is required to repress AP2L1 in the leaves and promote flowering. Interestingly, the levels of miR172 and both AP2L genes modulate the flowering response to different vernalization treatments in winter cultivars. In summary, our results show that conserved and grass specific gene networks interact to modulate the flowering response, and that natural or induced mutations in AP2L genes are useful tools for fine-tuning wheat flowering time in a changing environment.
Author summary
Reproductive success is essential for species survival, and in cultivated crops to maximize yield. Plants can sense and integrate different internal and environmental signals to ensure that flowering occurs under optimal conditions. In the temperate cereals, wheat and barley, specific mechanisms have evolved that guarantee flowering is promoted by the longer days of spring only after the plants have been exposed to the cold days of winter, a process called vernalization. In this work, we characterized the interactions between the vernalization requirement and a conserved pathway that integrates plant age into flowering regulation. This pathway involves the sequential action of two microRNAs, miR156 and miR172. In spring wheat cultivars, miR156 expression decreases with plant age, while miR172 expression increases. This results in the downregulation of its targets, the APETALA2-like (AP2L) flowering repressors, and the induction of flowering. In winter wheat cultivars, however, the induction of miR172 and the downregulation of AP2L1 is decoupled from miR156, and induction of the VERNALIZATION1 gene by vernalization is required to repress AP2L1 and promote flowering. Our results show that natural or induced mutations in the AP2L genes are useful tools for fine-tuning wheat flowering time in a changing environment.
Introduction
The precise control of flowering is central to plant reproductive success, and in cultivated cereals to maximize grain yield. Plants have evolved mechanisms that integrate various endogenous and environmental signals such as changes in day-length and temperature that enable them to flower under conditions that optimize seed production. Flowering takes place during a particular time of the year in response to perception of seasonal cues, such as changes in the length of the day or the night by a process called photoperiodism [1]. Temperate grasses, which include the agronomically important crops wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.), flower in the spring or early summer in response to shorter nights (longer photoperiods) and are referred to as long-day (LD) plants (e.g. [2,3]). Many plants adapted to temperate climates also have a winter annual or biennial life cycle strategy [4]. These plants become established in the fall, overwinter, and flower rapidly in the spring. Essential to this adaptive strategy is that flowering does not occur prior to winter, during which flowering would not lead to successful reproduction. Thus, these plants have evolved regulatory mechanisms to prevent fall flowering and sense the passing of winter to establish competence to flower [5]. The process by which flowering is promoted by a long exposure to cold temperatures is known as vernalization [5].
Comparisons of flowering regulatory networks in different plant species revealed several conserved genes and gene families, but also clade specific genes [6–8]. Different plant clades have evolved flowering network architectures that include novel components, extensive variation in gene expression patterns and/or interactions among conserved genes. Still, a common feature of these networks is that they converge on a small number of floral integrator genes that initiate the early stages of flowering [6].
In the temperate grasses, vernalization and photoperiod pathways converge in the transcriptional activation of FLOWERING LOCUS T (FT)-like genes in the leaves [1,9] and allelic variation at the FT1 locus in both barley and wheat is responsible for differences in heading time [9–11]. The central role of FT-like genes in initiating flowering appears to be conserved across flowering plants [2,12], and the timing of flowering depends largely on changes in FT expression in leaves. FT encodes a mobile protein that travels from the leaves to the shoot apical meristem (SAM) [13,14] where it interacts with the bZIP transcription factor FD to activate floral genes and transform the vegetative meristem into a floral meristem [15–18].
The major determinant of the photoperiodic regulation of FT1 in wheat and barley is PHOTOPERIOD1 (PPD1 or PRR37) [19]. PPD1 encodes a protein with a pseudo-receiver domain and a CONSTANS, CONSTANS-like, TIMING OF CAB EXPRESSION 1 (CCT) domain that promotes FT1 expression under LD conditions [19]. PPD1 is the result of a grass specific duplication event (PRR37/PPD1 and PRR73) that is independent of the PRR3-PRR7 duplication in Arabidopsis thaliana (Arabidopsis) [20]. The two Arabidopsis genes are part of the circadian clock, but in the temperate grasses PPD1 has a more specialized role in the photoperiod pathway [19,21]. PPD1 expression in the leaves is regulated by the circadian clock and phytochrome-mediated light signaling pathways, ensuring that PPD1 can only promote FT1 expression under LD conditions [3,19,21–24].
Mutations in the PPD-H1 coding region in barley result in non-functional or hypomorphic alleles with reduced ability to induce flowering under LD [19]. By contrast, deletions in the promoter regions of PPD-A1 (Ppd-A1a allele) and PPD-D1 (Ppd-D1a allele) homeologs in wheat result in increased PPD1 expression and accelerated heading under short day (SD) relative to the ancestral Ppd1b allele [25,26]. As a result, plants carrying the Ppd1a alleles show reduced photoperiod response and are designated as photoperiod insensitive (PI), whereas plants carrying the Ppd1b allele are designated as photoperiod sensitive (PS) [25,26]. In addition to PPD1, temperate grasses have a parallel photoperiod pathway that involves CONSTANS (CO)-like genes, CO1 and CO2 [22]. However, while in other species CO-like genes play a dominant role in photoperiod perception [6,12], in wheat and barley CO-like genes have a limited effect that is more relevant in the absence of PPD1 or when PPD1 has reduced function [22,27,28].
In wheat, FT1 expression is also regulated by the vernalization pathway. A current molecular model of vernalization in temperate grasses consists of three large-effect loci acting in a regulatory loop including VRN1, VRN2 and FT1 (also known as VRN3 in wheat, [9]). VRN2, which is a LD flowering repressor, antagonizes the role of PPD1 as a LD flowering promoter, preventing the induction of FT1 in the leaves during the fall [29]. VRN2 encodes a protein containing a zinc finger motif and a CCT domain and is orthologous to rice GHD7 [30,31] but has no known orthologs in eudicots [29,31]. Wheat and barley plants harboring loss-of-function VRN2 alleles display a spring growth habit [29,32–35]. VRN1 encodes a MADS box transcription factor of the SQUAMOSA clade [36,37], and its expression in leaves and apices is induced by vernalization [36]. Winter wheat varieties with a functional but recessive vrn1 allele require several weeks of vernalization to induce VRN1 and acquire competence to flower [36,38,39]. In contrast, spring wheat varieties carrying dominant Vrn1 alleles, which are expressed in the absence of cold temperature, bypass the vernalization requirement [38,40,41].
The expression of VRN1 in the leaves promotes the repression of VRN2 [41]. Thus, at the onset of spring as day length is extended, VRN2 expression levels are low due to the induction of VRN1 by the previous winter’s cold temperatures allowing PPD1 to induce the expression of FT1 in the leaves. In addition, a positive feedback loop between VRN1 and FT1 results in elevated levels of VRN1 in the presence of elevated FT1 levels and vice versa [42–44]. Therefore, in winter plants the repression of FT1 by VRN2 in the fall guarantees that promotion of flowering by long days is blocked until plants have been exposed to winter temperatures, and the positive feedback loop in the spring secures the commitment to flowering (reviewed in [43]).
In addition to the major photoperiodic and vernalization genes, additional genes and pathways integrate multiple signals that impact flowering, such as the circadian clock, the nutritional and developmental status of the plant and a variety of biotic and abiotic stressors [7,45–51]. For example, members of the APETALA2-like (AP2L) family (euAP2 lineage) of transcription factors have been shown to act as flowering repressors [52,53]. These transcription factors, defined by the presence of two AP2-like domains in tandem, are highly conserved in plants. They play important roles in the regulation of the flowering transition by integrating information related to the age of the plant and environmental signals [54].
In Arabidopsis, AP2L transcription factors repress FT transcription in the leaves as well as other flowering activator genes in the SAM of juvenile plants [55–57]. In addition, AP2L proteins interact with CO-like proteins and inhibit CO activity [57]. As a result, Arabidopsis plants with mutations in multiple AP2L members are rapid flowering under both LD and SD conditions [56,58,59]. Interestingly, in the perennial species Arabis alpina (A. alpina), a close relative of Arabidopsis, the AP2L gene PERPETUAL FLOWERING2 (PEP2) is a flowering repressor that controls the vernalization response and contributes to the perennial life cycle [60,61]. The function of AP2L genes as flowering repressors is also conserved in monocot species. In maize (Zea mays), overexpression of the AP2L genes Glossy15 and ZmTARGET OF EAT1 (ZmTOE1 or Related to APETALA2.7, ZmRAP2.7) delays flowering [62,63]. In rice (Oryza sativa), the AP2L genes INDETERMINATE SPIKELET 1 (OsIDS1), SUPERNUMERARY BRACT (SNB) and OsTOE1 also act as flowering repressors [64,65]. In wheat, loss-of-function mutations in the domestication gene Q (also named AP2L5 = wheat ortholog of IDS1) accelerate flowering [66,67].
The expression of AP2L genes is regulated at the post-transcriptional level by miR172, which is an ancient and conserved miRNA in plants [54]. Across flowering plant diversification, the expression of miR172 increases during and after the transition to flowering [58,59,62,64,68–71]. In reproductive tissues, miR172 controls AP2L expression to regulate inflorescence and flower development [72,73]. In wheat inflorescences, this regulation is important to control spikelet density, floret number, and the free-threshing character of the spike in domesticated wheat [66,74].
The repression of AP2L genes by miR172 is also important to control the timing of the flowering transition. Ectopic expression of MIR172 genes phenocopies the rapid flowering of mutants in multiple AP2L genes in Arabidopsis, rice and wheat [56,58,64–66]. On the other hand, plants with reduced activity of miR172, including Arabidopsis CRISPR mutants in multiple miR172 loci [70,71], plants expressing artificial target mimics against miR172 (henceforth MIM172) or miR172-resistant versions of AP2L genes [56,64–66,75], all display a delayed flowering phenotype.
The expression of miR172 in vegetative tissues is controlled by the environment and the developmental status of the plant [58,59,62,64,68,76–78]. As plants age, the expression levels of miR172 increase. This is part of a conserved network that involves another conserved miRNA named miR156, which controls the juvenile to adult vegetative phase transition and flowering competence in several species [76,78]. In this so-called plant age pathway, miR156 is expressed at high levels in juvenile stages and represses the expression of a group of SQUAMOSA-PROMOTER-BINDING-like (SPL) genes [79]. As plants grow and in response to the increase in sugar levels [80,81], miR156 expression goes down resulting in an increased expression of SPLs, which in turn activate the expression of miR172 in the leaves. The increased levels of miR172 result in repression of AP2L genes expression, which promotes adult character traits in leaves and also flowering competence [78,82,83].
Our previous studies focused on the role of miR172 and AP2L genes in wheat spike development [66,84]. In this work, we focus on the role of the miR172-AP2L module in the regulation of the flowering transition through its interaction with the temperate grasses specific flowering pathway involving the VRN1-VRN2-FT1 genetic feedback loop. Using a combination of mutants, transgenics, and different growth conditions, we show that miR172 promotes the flowering transition in wheat, while its targets AP2L genes work as repressors of this transition. In addition, we show that miR172 and AP2L genes regulate the expression of VRN1-VRN2-FT1 genes in leaves and modulate the flowering response in spring and winter wheats under different environmental conditions. Finally, we describe mutations in these genes that could be useful tools to fine-tune wheat flowering time in a changing environment.
Results
miR172 promotes the flowering transition in spring wheat
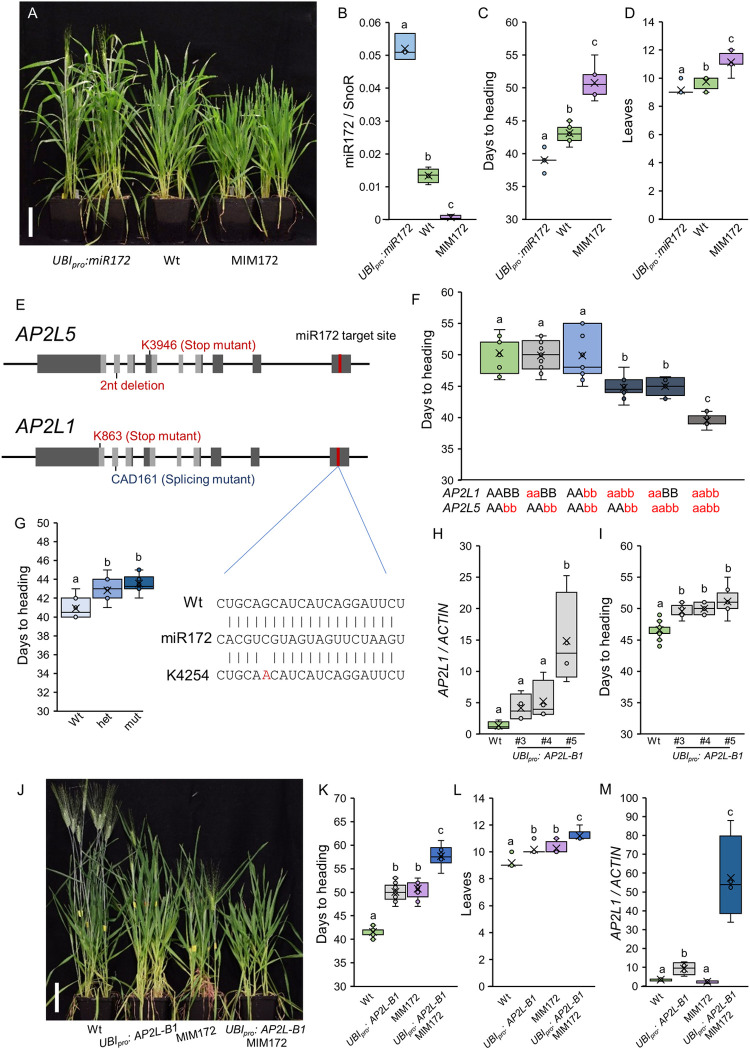
To study the role of miR172 in the control of the flowering transition in wheat, we first analyzed transgenic lines with altered levels of miR172 that were previously generated in the tetraploid wheat variety Kronos (Triticum turgidum subsp. durum) [66] (Fig 1A). Kronos has a spring growth habit determined by the Vrn-A1c allele (deletion in intron 1), and a reduced photoperiod response conferred by the Ppd-A1a allele [41]. We selected and compared independent T2 transgenic lines overexpressing either miR172 (UBIpro:miR172) or a target mimic against miR172 (MIM172). Quantification of miR172 levels by qRT-PCR in a fully expanded fifth leaf confirmed a 4-fold increase in miR172 expression levels in UBIpro:miR172 plants compared to wild type, and a 20-fold reduction in MIM172 transgenic plants (Fig 1B and Data A in S1 Data).
Fig 1. miR172 promotes flowering in wheat by repressing AP2L genes.
(A) Six-week-old wild type Kronos plants (Wt) compared to UBIpro:miR172 and MIM172 transgenic plants in the same genetic background grown under LD. Scale bar = 10 cm. (B) Box plots showing miR172 expression levels in the 5th leaf of UBIpro:miR172, wild type Kronos (Wt) and MIM172 plants grown under LD. Mature miR172 levels were determined by qRT-PCR using the small nucleolar RNA 101 (SnoR) as internal reference. Data correspond to four independent biological replicates. (C-D) Box plots showing days to heading (C; n ≥ 7) and number of leaves produced by the main tiller (D; n ≥ 7) in the same genotypes and conditions as in B. (E) Schematic diagrams showing the gene structures of AP2L5 and AP2L1. Exons are indicated as dark grey boxes, AP2 domains are indicated in light grey, and the miR172-target sites in red. The positions of the TILLING mutations K3946 (AP2L5) and K863 (AP2L1) in the A homeologs are indicate above the gene structures, and the natural variant with 2 nucleotides deletion (AP2L5) and CAD161 splicing mutant (AP2L1) in the B homeologs below. (F) Box plots showing days to heading under LD conditions for wild type (Wt), plants harboring mutations in only one AP2L1 homeolog (aaBB and AAbb), in both AP2L1 homeologs (ap2l1-null = aabb), ap2l5-null mutants, and apl2l1 ap2l5 combined null mutants (n ≥ 8). (G) Box plots showing days to heading under LD conditions for an F2 population segregating for the K4254 mutation in the miR172-target site of AP2L-A1 gene (n ≥ 10).; Wt = homozygous wild type plants, mut = homozygous K4254 mutants, het = heterozygous plants. The interaction between the wild type miR172-target site and miR172 is shown in the right. The TILLING line K4254 has a G>A mutation in the miR172 target site that reduces the free energy of interaction with miR172. (H-I) Box plots showing the expression levels of AP2L1 in the 5th leaves (H; n = 4) and days to heading (I; n ≥ 5) for wild type Kronos (Wt) and three independent UBIpro:AP2L-B1 transgenic lines grown under LD. (J-M) Wild type Kronos (Wt), UBIpro:AP2L1 (transgenic line #5), MIM172 and MIM172 UBIpro:AP2L-B1#5 plants grown under LD. (J) Eight-week-old plants. Scale bar = 10 cm. (K-M) Box plots showing days to heading (K; n ≥ 6), number of leaves produced by the main tiller (L; n ≥ 5) and expression levels of AP2L1 in the 5th leaves (M). Different letters above the box plots indicate significant differences based on Tukey tests (P < 0.05), except for panels (D), (F) and (L) where non-parametric Kruskal-Wallis tests were used (Data A in S1 Data).
Under LD conditions (16 h light / 8 h dark), plants overexpressing miR172 headed 4 days earlier (Fig 1C) and produced 0.6 fewer leaves than the wild type (Fig 1D), whereas MIM172 plants headed 8 days later and produced 1.4 more leaves than the wild type (Fig 1C and 1D). Under SD conditions (8 h light / 16h dark) Kronos plants headed in approximately 80 days (S1A and S1B Fig). UBIpro:miR172 plants flowered earlier than the wild type, and MIM172 plants flowered later than the wild type, similarly to the LD conditions. However, the differences in heading time between UBIpro:miR172 and MIM172 plants were larger under SD (40 days and 5 leaves, S1A–S1C Fig) than under LD (12 days and two leaves, Fig 1C and 1D). These results show that in spring wheat, miR172 accelerates the transition to flowering both under LD and SD conditions.
Mutations in AP2L genes lead to an acceleration of flowering
We next explored the role of AP2L transcription factors, which are known targets of miR172 in the regulation of flowering time [54]. In wheat, we previously identified four AP2L genes with miR172 target sequences (named AP2L1, AP2L2, AP2L5, and AP2L7; [66,84]; S1 Table). Loss-of-function AP2L5 mutants (Fig 1E) showed an early flowering phenotype [66,84] (Fig 1F), whereas the heading time of a null mutant for AP2L2 was not different to the wild type control [84]. We identified another AP2L gene (named AP2L1), which is orthologous with TOE1 (TARGET OF EARLY ACTIVATION TAGGED (EAT) 1), a known flowering repressor in Arabidopsis and maize [58,63,84]. Therefore, in this work we further characterized the function of AP2L1. We identified TILLING lines with loss-of-function mutations in both the A (K863) and B (CAD0161) homeologs and crossed them to generate an ap2l1-null mutant in Kronos (Fig 1E, see Material and Methods). Under LD conditions, the ap2l1-null plants headed 5 days earlier than both wild type control and lines harboring mutations in only one AP2L1 homeolog (Fig 1F). The acceleration of flowering was similar to an ap2l5-null mutant grown in the same chamber. We also generated and tested ap2l5 ap2l1 combined null mutant plants in the same growing conditions. The combined null mutant headed significantly earlier than each of the single gene null mutants and 10 days earlier than the wild type (Fig 1F). These results suggest overlapping and additive roles for these two miR172-targets in the control of flowering time in spring wheat. The ap2l1-null mutant did not show any of the spikelet or floret phenotypes previously described for the ap2l5-null mutant [66,84] (S2 Fig).
miR172 promotes flowering by repressing AP2L genes
To study the effect of miR172 on AP2L1 and AP2L5, we first checked the expression of these genes in the fifth leaves of Kronos, MIM172 and UBIpro:miR172 plants, using the same leaf samples that showed highly significant differences in miR172 expression in Fig 1B. Interestingly, we did not observe differences in the expression of AP2L1 between MIM172, wild type and UBIpro:miR172 samples (S3A Fig), and the expression of AP2L5 was higher in MIM172 compared to wild type but no significant differences were observed between wild type and UBIpro:miR172 (S3B Fig and Data I in S1 Data). Similar observations were described in other species, where steady-state transcript levels of AP2L genes remained constant upon changes in miR172 levels [58,72,85]. Those observations suggested that miR172 could control AP2L expression at the translational level [58,72] or that the AP2L proteins could regulate their own transcript levels through feedback loops [55,85].
We further characterized the role of miR172 and AP2L1 in flowering by studying a Kronos TILLING line (K4254) with a point mutation in the miR172 target site of the AP2L-A1 homeolog (Fig 1G). This mutation lies in the same position within the miR172 target site as the mutation that generates the dominant Tasselseed6 allele of IDS1 in maize [68] and is expected to reduce the interaction with miR172. We backcrossed K4254 two times with wild type Kronos and analyzed heading time under LD in a BC1F2 segregating population. Plants carrying this dominant mutation headed 3.25 days later than the sister wild type plants (Fig 1G and Data A in S1 Data). This result is consistent with AP2L1 being a flowering repressor targeted by miR172.
We confirmed the flowering repression activity of AP2L1 using transgenic plants constitutively expressing the wild type coding region of the AP2L-B1 gene under the maize UBIQUITIN promoter (Fig 1H). Three independent UBIpro:AP2L-B1 lines showed significant delays in heading time compared to the non-transgenic sister lines (2.9–4.5 days, Fig 1I). We then selected the line UBIpro:AP2L-B1#5 with the highest AP2L1 transcript levels (Fig 1H) and crossed it with MIM172, which has reduced miR172. In the resulting F2 population grown under LD conditions (Fig 1J), plants harboring UBIpro:AP2L-B1#5 or MIM172 alone headed later and produced more leaves than the wild type (Fig 1K and 1L). Plants carrying both transgenes (UBIpro:AP2L-B1 and MIM172) showed a larger delay in heading (16.1 days) and produced two more leaves on the main tiller compared to the wild type control, and were significantly later than the individual transgenic lines (Fig 1K and 1L).
Analysis of AP2L1 expression levels in these lines showed that plants carrying only the MIM172 transgene did not show differences in AP2L1 expression compared to wild type (as observed in S3A Fig), but plants harboring both transgenes have the highest AP2L1 transcript levels (Fig 1M).We hypothesize that the expression of AP2L-B1 under the constitutive UBIQUITIN promoter likely disrupted the putative feedback loop of the AP2L1 protein on its own expression, and revealed the activity of miR172 on the AP2L1 transcript levels. Taken together, these results support the role of miR172 as a repressor of AP2L1 and a promoter of flowering.
miR172 and AP2L genes regulate flowering by modulating the expression of VRN1, VRN2 and FT1 in the leaves
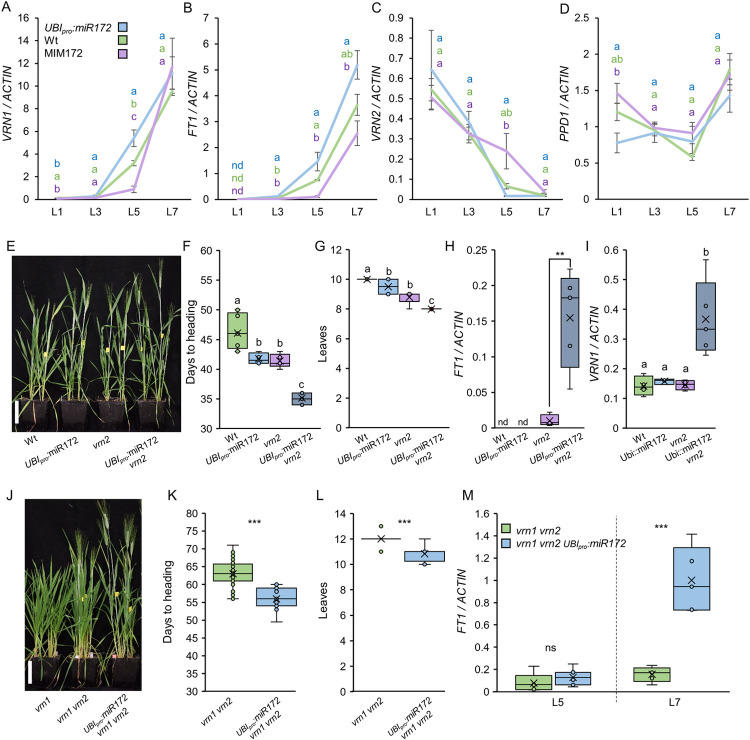
We then studied how miR172 and AP2L interact with the wheat flowering network involving PPD1 and the VRN1-VRN2-FT1 regulatory loop. To this end, we performed a time-course expression analysis using leaves collected from Kronos wild type, UBIpro:miR172 and MIM172 transgenic plants at different ages grown under LD. In the three different genotypes, the expression of flowering promoters VRN1 and FT1 (Fig 2A and 2B) increased with plant age, from juvenile leaf 1 (L1) to adult leaf 7 (L7), while the expression of the flowering repressor VRN2 decreased as plants approached flowering (Fig 2C). In the fifth leaf, we observed significantly higher expression of VRN1 (Fig 2A) and FT1 (Fig 2B) in the early flowering UBIpro:miR172 plants than in the wild type. By contrast, in the same leaf, we observed significantly lower expression of VRN1 (Fig 2A) and FT1 (Fig 2B) in the late flowering MIM172 plants than in the wild type. A similar expression pattern was observed for the two VRN1-related SQUAMOSA MADS-box genes FUL2 and FUL3, whose expression was higher in the fifth leaf of UBIpro:miR172 and lower in MIM172. For these two genes, the differences between wild type and MIM172 were not significant in L5, but were significant in L7 (S4 Fig and Data J in S1 Data). In contrast, the expression levels of PPD1 were similar among leaves of different ages, and no significant differences with the wild type were observed for UBIpro:miR172 or MIM172 (Fig 2D).
Fig 2. miR172 controls flowering through the regulation of FT1 expression in leaves.
(A-D) Expression levels of VRN1 (A), FT1 (B), VRN2 (C), and PPD1 (D) determined by qRT-PCR in the 1st (L1), 3rd (L3), 5th (L5) and 7th (L7) leaves of UBIpro:miR172, wild type Kronos (Wt) and MIM172 plants grown under LD. ACTIN was used as internal reference. Data correspond to four independent biological replicates. (E-I) Population segregating for wild type Kronos (Wt), UBIpro:miR172, vrn2 and UBIpro:miR172 vrn2 plants grown under LD. (E) Six-week-old plants. Scale bar = 10 cm. (F-G) Box plots showing days to heading (F; n ≥ 4) and total number of leaves produced by the main tiller (G; n ≥ 4). (H-I) Box plots showing FT1 (H) and VRN1 (I) expression levels determined by qRT-PCR in the first leaves. ACTIN was used as internal reference. Data correspond to four independent biological replicates. (J) Nine-week-old vrn1 null, vrn1 vrn2 combined mutant and vrn1 vrn2 UBIpro:miR172 plants growing under LD. Scale bar = 10 cm. (K-M) vrn1 vrn2 combined mutant and vrn1 vrn2 UBIpro:miR172 plants grown under LD. (K-L) Box plots showing days to heading (K; n ≥ 11) and number of leaves produced by the main tiller (L; n ≥ 12). (M) Box plots showing FT1 expression levels determined by qRT-PCR in the 5th (L5) and 7th (L7) leaves. ACTIN was used as internal reference. Data correspond to five independent biological replicates. Different letters above the box plots indicate significant differences based on Tukey tests (P < 0.05) in panels (A-D), (F) and (I), on Kruskal-Wallis non-parametric testes in (G) and (L), and on t-tests in panels (H), (K) and (M) (Data B in S1 Data). ns = not significant, * = P < 0.05, ** = P < 0.01, *** = P < 0.001. The color of the letters in A-D correspond to the color of the genotypes.
We also checked the expression of these genes in the third and sixth leaves of plants grown under SD conditions (S1D–S1G Fig). The results were similar to LD, with UBIpro:miR172 plants expressing significantly higher levels of FT1 and VRN1 in both the third and sixth leaves, lower levels of VRN2 (only L6), and no significant changes in PPD1 expression compared to wild type Kronos (Data H in S1 Data). The MIM172 transgenic plants showed similar levels of VRN2 compared to wild type in both leaves, and lower levels of VRN1 and FT1 in the sixth leaf, although the differences were not significant (S1D–S1G Fig). Taken together, these results indicate that constitutive expression of miR172 activity in leaves under both LD and SD conditions is associated with increased levels of the flowering promoters VRN1 and FT1, and decreased levels of the flowering repressor VRN2.
We also analyzed the expression of VRN1, VRN2, and FT1 in the fifth leaf of plants from an F2 population segregating for UBIpro:AP2L-B1 and MIM172 grown under LD (S5 Fig). In agreement with their late flowering phenotype, the UBIpro:AP2L-B1 MIM172 plants showed the largest reduction in VRN1 and FT1 expression compared with the wild type (S5A and S5B Fig and Data K in S1 Data), while the expression of the flowering repressor VRN2 was significantly increased (S5C Fig). The transgenic plants with either UBIpro:AP2L-B1 or MIM172 showed intermediate values for all three genes. These results indicate that AP2L genes repress wheat flowering likely by controlling the expression of these three central wheat flowering genes.
miR172 regulates FT1 expression in the absence of VRN1 and VRN2
From the previous experiments, it was not possible to determine which of the three genes is affected by the miR172-AP2L module because VRN1-VRN2-FT1 are interconnected through feedback regulatory loops. To answer this question, we first crossed the UBIpro:miR172 line with vrn2 loss-of-function Kronos mutant plants [32] and analyzed the flowering phenotype in the resulting F2 population. Under LD conditions, UBIpro:miR172 lines phenocopy the rapid flowering of the vrn2 mutant, both in days to heading and leaf number (Fig 2E–2G). Moreover, the UBIpro:miR172 vrn2 plants showed an additive early phenotype as compared to the single UBIpro:miR172 or vrn2 plants (Fig 2E–2G). We collected tissue from the first leaves of these plants to check early responses in the expression of flowering genes. We were not able to amplify FT1 transcripts in wild type or UBIpro:miR172 first leaf samples, but we detected a low FT1 expression level in vrn2 mutants (Fig 2H). Interestingly, UBIpro:miR172 vrn2 showed a significant increase in FT1 expression compared to the vrn2 mutant (Fig 2H). VRN1 expression was detected in all lines and no difference was observed between wild type, UBIpro:miR172 and vrn2 samples. However, VRN1 expression was significantly higher in UBIpro:miR172 vrn2 plants (Fig 2I). These results indicate that overexpression of miR172 can promote FT1 and VRN1 expression in the absence of VRN2.
Since FT1 and VRN1 can also regulate each other in a positive feedback loop [9,17,42,50,86], we also introduced the UBIpro:miR172 transgene into a vrn1 vrn2 mutant background [41] to further dissect these interactions. Plants with loss-of-function mutations in VRN1 (vrn1) cannot repress VRN2 expression and have very delayed heading [41] (Fig 2J). Elimination of VRN2 in the vrn1 vrn2 mutant background accelerates the flowering transition [41] (Fig 2J). Interestingly, the UBIpro:miR172 vrn1 vrn2 plants headed on average 8 days earlier than vrn1 vrn2 and produced one fewer leaf (Fig 2J–2L), indicating that UBIpro:miR172 can promote flowering in the absence of VRN1 and VRN2. We collected tissue from the fifth and seventh leaves of these plants and measured FT1 mRNA levels. In agreement with the rapid flowering, we observed a significant increase in FT1 expression in seventh leaves of UBIpro:miR172 vrn1 vrn2 compared to vrn1 vrn2 plants (Fig 2M). Taken together, these results indicate that miR172 can promote flowering through the regulation of FT1 expression in leaves independently of VRN1 and VRN2.
miR156 modulates wheat flowering time through regulation of miR172 and AP2L1 expression
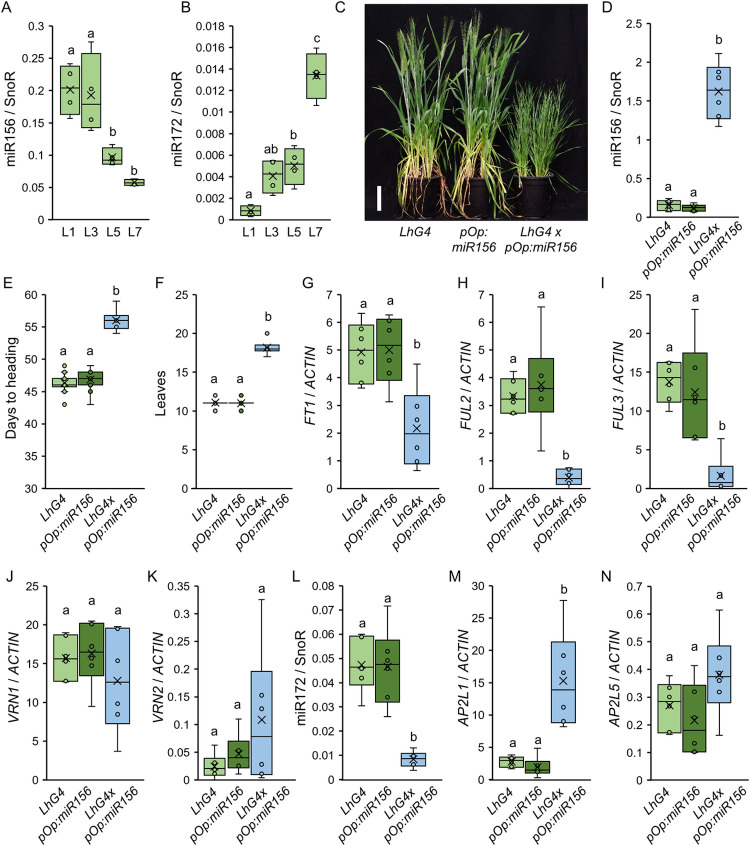
Next, we examined the connection between the miR172-AP2L module described above and the developmentally regulated miR156. A time-course experiment in leaves of wild type Kronos grown under LD conditions showed higher levels of miR156 in the first and third leaves followed by downregulation in adult fifth and seventh leaves (Fig 3A). In the same samples, mature miR172 levels increased with plant age, following an opposite trend to miR156 (Fig 3B). These patterns are similar to those observed in other species [76,78,83], suggesting that the age-related miR156-miR172 pathway is conserved in wheat.
Fig 3. Overexpression of miR156 delays the flowering transition in wheat.
(A-B) Box plots showing the expression levels of miR156 (A) and miR172 (B) determined by qRT-PCR in the 1st (L1), 3rd (L3), 5th (L5) and 7th (L7) leaves of wild type Kronos plants growing under LD. SnoR was used as an internal reference. Data correspond to four independent biological replicates. (C-N) UBIpro:LhG4 (LhG4), pOp:miR156 and UBIpro:LhG4/pOp:miR156 plants growing under LD. (C) Six-week-old plants. Scale bar = 10 cm. (D) Box plots showing miR156 expression levels determined by qRT-PCR in the 9th leaf. Data correspond to six independent biological replicates. (E-F) Box plots showing days to heading (E; n ≥ 6) and the number of leaves produced by the main tiller before the transition to the spike (F; n ≥ 6). (G-N) Box plots showing the expression levels of FT1 (G), FUL2 (H), FUL3 (I), VRN1 (J), VRN2 (K), miR172 (L), AP2L1 (M) and AP2L5 (N) determined by qRT-PCR in the 9th leaf. ACTIN was used as internal reference for FT1, FUL2, FUL3, VRN1, VRN2, AP2L1 and AP2L5, and SnoR was used for miR172. Data correspond to six independent biological replicates. Different letters indicate significant differences in Tukey tests (P < 0.05), except for panel (F) where non-parametric Kruskal-Wallis pair-wise tests were used (Data C in S1 Data).
To investigate the role of miR156 on the regulation of the miR172-AP2L module in wheat, we first generated transgenic plants expressing constitutively high levels of miR156. We identified 5 loci per genome in tetraploid wheat that encode miR156 precursor sequences that can be potentially processed to generate mature miR156 sequences (S2 Table). We decided to clone and overexpress the miR156b,c locus, which has been studied in several monocot species [87]. Interestingly, the miR156b,c loci from rice, maize and Brachypodium include two precursors that are expressed in tandem, while Triticeae species contain an extra precursor so the wheat miR156b,c locus includes three precursors in tandem (S6A Fig).
Overexpression of the B genome miR156b,c locus in tetraploid Kronos using the maize UBIQUITIN promoter resulted in T0 plants that produced many tillers and had a bushy appearance (S6B Fig), as previously reported [88]. However, these plants were sterile and did not set grain. To generate stable transgenic lines overexpressing miR156, we used the LhG4/pOp two-component system ([89]; see details in Material and Methods). In this system, miR156 was only overexpressed after crossing plants harboring UBIpro:LhG4 with those containing the pOp:miR156 construct (Fig 3C). Compared with F1 plants harboring only one of the constructs, the F1 UBIpro:LhG4/pOp:miR156 plants expressed high levels of miR156 (Fig 3D), required 10 more days to head (Fig 3E) and produced on average 8 more leaves than the controls (Fig 3F). The delayed flowering of the UBIpro:LhG4/pOp:miR156 plants correlated with a significant reduction in FT1, FUL2, and FUL3 expression (Fig 3G–3I), a significant reduction in mature miR172 expression (Fig 3L), and higher levels of AP2L1 (Fig 3M) compared with the controls. The expression levels of VRN1, VRN2 and AP2L5 were not affected by the overexpression of miR156 (Fig 3J, 3K, and 3N).
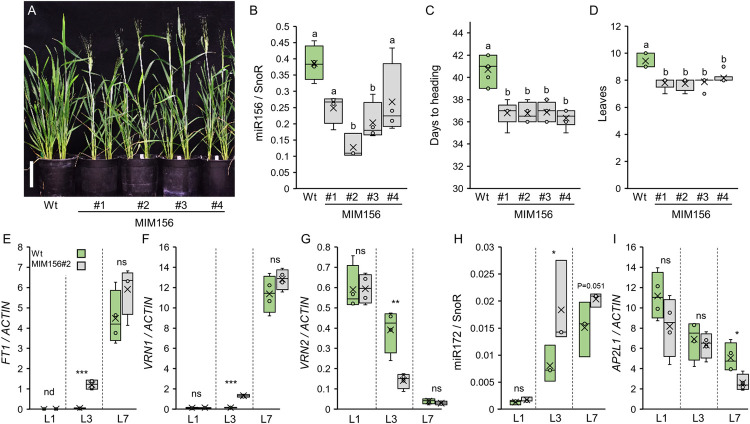
To complement the analysis of the transgenic lines overexpressing miR156, we generated transgenic lines expressing a target mimic to reduce miR156 activity in vivo (MIM156) (Fig 4A). We generated four independent MIM156 lines in Kronos that had lower levels of miR156 (Fig 4B) and characterized their flowering phenotypes under LD conditions. The MIM156 plants headed 4 days earlier (Fig 4C) and produced 1–2 fewer leaves than the non-transgenic sister lines (Fig 4D), indicating an early transition to reproductive development. We then checked the expression of flowering genes in the first, third and seventh leaves of wild type and MIM156#2 plants grown under LD conditions. Consistent with their early flowering, the third leaves of MIM156#2 plants expressed higher levels of FT1 (Fig 4E) and VRN1 (Fig 4F), and lower levels of VRN2 (Fig 4G).
Fig 4. Transgenic target mimic MIM156 induces expression of flowering promoting genes and accelerates heading time.
(A) Five-week-old wild type Kronos (Wt) and T1 plants of four independent MIM156 transgenic lines grown under LD. Scale bar = 10 cm. (B) Box plots showing the expression levels of miR156 determined by qRT-PCR in the 1st leaves (L1) of wild type Kronos plants (Wt) and T1 plants of four independent MIM156 lines grown under LD. SnoR was used as internal reference. Data correspond to four independent biological replicates. (C-D) Box plots showing days to heading (C; n ≥ 4) and the number of leaves produced by the main tiller (D; n ≥ 4) for wild type Kronos (Wt) and four independent MIM156 T1 lines grown under LD. (E-I) Box plots showing the expression levels of FT1 (E), VRN1 (F), VRN2 (G), miR172 (H) and AP2L1 (I) determined by qRT-PCR in the 1st (L1), 3rd (L3) and 7th (L7) leaves of wild type Kronos (Wt) and MIM156#2 growing under LD. As internal references, we used ACTIN for FT1, VRN1, VRN2 and AP2L1, and SnoR for miR172. Data correspond to four independent biological replicates. Different letters above the box plots indicate significant differences based on Tukey tests in panels B and D and Kruskal-Wallis tests in panel C (P < 0.05). Differences in panels E-I are based on t-tests (Data D in S1 Data). ns = not significant, * = P ≤ 0.05, ** = P ≤ 0.01, *** P ≤ 0.001.
We hypothesized that part of the rapid flowering phenotype of MIM156 plants was mediated by the induction of miR172. In agreement with this hypothesis, we observed higher levels of miR172 in the third and seventh leaves of MIM156 plants (Fig 4H), and a reduction in AP2L1 expression in the seventh leaf (Fig 4I). In addition, we crossed MIM156 and MIM172 transgenic plants and analyzed the flowering phenotype of the F2 population. As expected, the MIM156 MIM172 plants were later and produced more leaves than single MIM156 plants (S7A and S7B Fig and Data L in S1 Data). However, MIM156 could still accelerate flowering in the presence of MIM172 when compared to single MIM172 (S7A and S7B Fig).
Taken together, these results show that miR156 delays flowering in wheat, in part through the regulation of miR172 and AP2L1 expression.
Repression of AP2L1 is decoupled from the age-controlled downregulation of miR156 in winter wheat plants
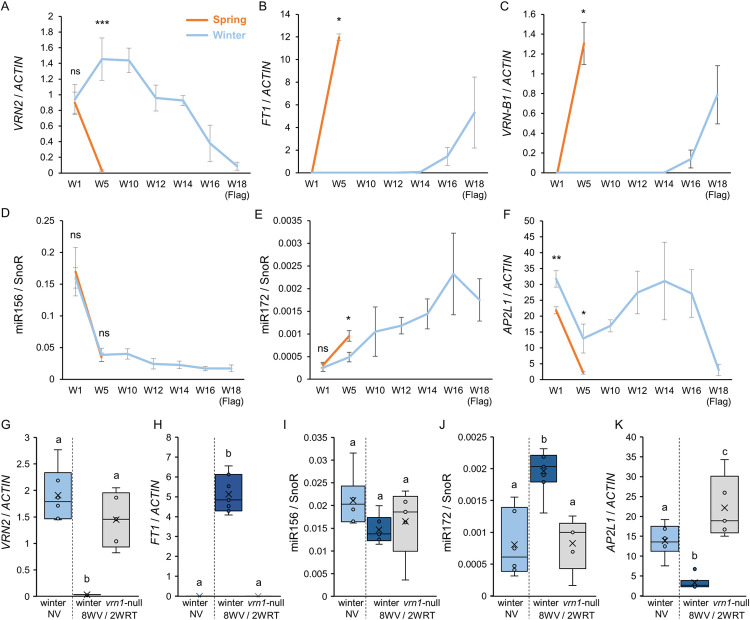
The previous results showed that miR156 levels affect the expression of miR172 and AP2L1 in leaves, which in turn modulates the flowering transition in spring wheat cultivars. However, in spring cultivars the flowering transition occurs rapidly, and it is difficult to distinguish the individual effects of plant age (miR156) and flowering induction on the miR172-AP2Ls module. Therefore, we studied the miR156-miR172 pathway during the flowering transition in winter wheat plants, which require a long exposure to cold temperatures in order to cause rapid flowering. To this end, we used a Kronos TILLING line that has a loss-of-function mutation in the spring VRN-A1 allele and a functional winter vrn-B1 allele, and therefore these plants have a winter growth habit ([41], henceforth “winter Kronos”). These lines show accelerated heading time and reduced leaf number with increased vernalization treatments, a response that is saturated after 6 weeks of vernalization (S8A and S8B Fig and Data M in S1 Data).
First, we performed a detailed time-course expression analysis in leaves collected from spring and winter Kronos plants of different ages grown under LD in the absence of cold. Under this condition, winter plants required ~100 days more than spring sister lines to head (S9A and S9B Fig and Data N in S1 Data). In spring plants, the expression of VRN2 was significantly downregulated while FT1 and VRN-B1 transcript levels were induced in the seventh leaf (L7; W5 = 5-week-old plants) compared to the first leaf (L1; W1 = 1-week-old plants), in agreement with their early flowering (Fig 5A–5C). In contrast, the expression of VRN2 remained high in 5 to 10-week-old winter plants, and then slowly declined until reaching very low levels in 18-week-old plants, which had already developed the flag leaf (Fig 5A). The downregulation of VRN2 was accompanied by an induction of FT1 and VRN-B1 between 14 and 18 weeks (Fig 5B and 5C).
Fig 5. Effect of vernalization and VRN1 expression on the transcript levels of flowering genes, miR156 and miR172 in spring and winter wheat.
(A-F) Time-course expression levels determined by qRT-PCR in leaves of spring (orange) and winter (blue) Kronos plants grown under LD conditions in the absence of a vernalization treatment. W1 = week 1, 1st leaf; W5 = week 5, 7th leaf; W10 = week 10, leaves 10th-11th; W12 = week 12, leaves 13th-14th; W14 = week 14, leaves 15th-16th; W16 = week 16, leaves 18th-19th; W18, week 18, flag leaves (20th-23rd). In both genotypes the fully expanded 7th leaves were collected during the 5th week. However, leaf samples were not collected on the same day (34 days for spring and 36–38 days after planting for winter). ACTIN was used as internal reference for (A) VRN2, (B) FT1, (C) VRN1, and (F) AP2L1, and SnoR for (D) miR156 and (E) miR172. * = P < 0.05, ** = P < 0.01, and *** = P < 0.001 in t-tests (A, D-F) and non-parametric Kruskal-Wallis tests (B and C) comparing expression at the same leaf in spring and winter Kronos plants. (G-K) Box plots showing the expression levels of VRN2 (G), FT1 (H), miR156 (I), miR172 (J) and AP2L1 (K) determined by qRT-PCR in the 7th leaf of winter Kronos plants without vernalization, and the 7th leaves of winter Kronos and vrn1-null mutants that were vernalized for 8 weeks and then moved to room temperature for two weeks. Samples from the 7th leaf were collected when the leaves were fully expanded. ACTIN was used as internal references for VRN2, FT1 and AP2L1, and SnoR for miR156 and miR172. Data correspond to at least 5 independent biological replicates. Different letters above the box plots in panels G-K indicate significant differences (P < 0.05) in Tukey tests, except for pane (H), where a non-parametric Kruskal-Wallis test was used (Data E in S1 Data).
We observed that mature miR156 was expressed at similar levels in juvenile leaves of spring and winter plants (W1), and its expression was then repressed to similar levels in the seventh leaves (W5) of plants with both growth habits (Fig 5D). Interestingly, the miR156 expression continued to show a gradual downregulation in later leaves of winter plants.
Between W1 and W5, miR172 levels increased in both spring and winter Kronos, but reached significantly higher levels in the spring lines (which were close to heading) than in the winter lines (Fig 5E). By contrast, the expression level of AP2L1 was significantly higher in the seventh leaves of winter plants compared with spring plants at W5 (Fig 5F).
In winter lines, we observed a progressive increase in the expression of miR172 with age until week 16 (Fig 5E). Interestingly, we did not observe a parallel downregulation of the transcript levels of AP2L1 (Fig 5F) and AP2L5 (S9C Fig) after W5. This result is similar to the limited effect of UBIpro:miR172 on AP2Ls transcript levels described in S2A and S2B Fig. After W14, we observed an abrupt downregulation of AP2L1 and AP2L5 that coincided with the upregulation of FT1 (Fig 5B), and the SQUAMOSA genes VRN1 (Fig 5C), FUL2 and FUL3 (S9D and S9E Fig).
For all the genes in Figs 5A–5F (Data E in S1 Data) and S9D and S9E Fig (Data N in S1 Data), the ANOVAS for the differences in gene expression along development were significant for both spring and winter lines, confirming that all these genes are developmentally regulated. Only AP2L5 (S9C Fig) showed no significant differences for the spring line and only marginally significant differences for the winter lines (P = 0.05). In addition, no significant differences in gene expression were detected between the spring line at W5 and the winter line at W18 in Figs 5A–5F (Data E in S1 Data) and S9D-E (Data N in S1 Data), suggesting a similar regulation of these genes at heading time in the two genotypes.
The downregulation of AP2L1 associated with the upregulation of SQUAMOSA MADS-box genes correlates with the negative regulation of AP2L genes by SQUAMOSA MADS-box genes reported in Arabidopsis [70,90]. A similar regulation may also explain the rapid repression of AP2L1 (and induction of miR172) observed at W5 in the spring Kronos lines. To further explore this hypothesis, we studied the expression of AP2L1 and miR172 in vernalized winter Kronos and vrn1-null mutant plants [41]. We vernalized 2-week-old winter plants for 8 weeks and collected L7 samples from non-vernalized and vernalized winter plants two weeks after the plants were taken out of the cold. The expression of VRN2 remained repressed in the vernalized winter plants 2 weeks after returning to room temperature but not in the vrn1-null mutant, consistent with previous findings [41] (Fig 5G). Expression of FT1 (Fig 5H), VRN-B1, FUL2 and FUL3 (S9G–S9I Fig) was induced only in vernalized winter plants with the functional VRN1 allele. The expression of miR156 was not modified by the vernalization treatment (Fig 5I), but we observed an induction of miR172 expression in the vernalized winter plants compared with non-vernalized winter and vernalized vrn1-null plants (Fig 5J). Moreover, the expression of AP2L1 was significantly downregulated in winter plants compared with the vrn1-null mutants after both lines were returned to room temperature from the vernalization treatment (Fig 5K).
Taken together, these results indicate that in the leaves of winter plants, the downregulation of miR156 expression with plant age is decoupled from the downregulation of AP2L genes expression, and that the induction of VRN1 by vernalization promotes the upregulation of miR172 and the repression of AP2L1.
miR172 modulates flowering time in winter wheat plants exposed to different vernalization treatments
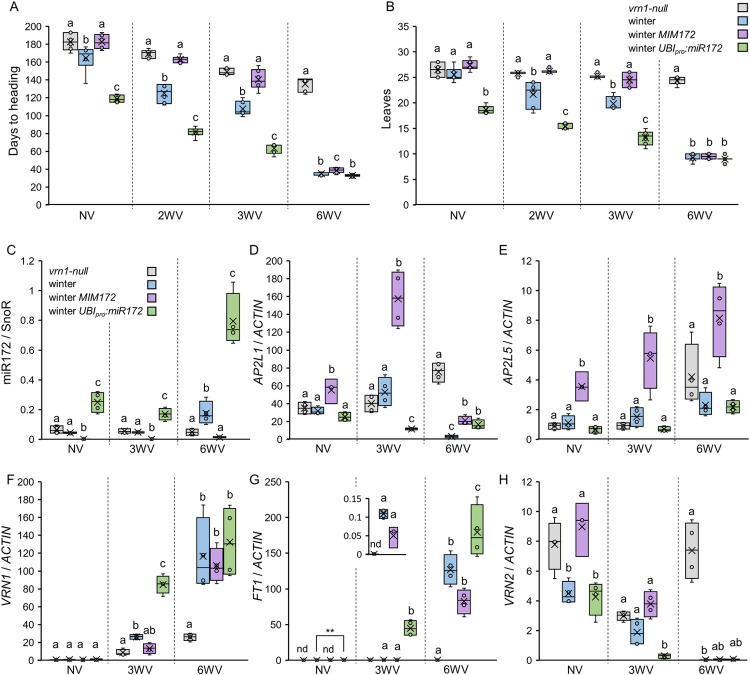
We next explored the role that the miR172-AP2L module has in the flowering transition of winter wheat plants. To this end, we transferred the UBIpro:miR172 and MIM172 transgenes into the winter Kronos background, and evaluated the flowering time of plants grown under LD conditions without vernalization, or of those vernalized for 2, 3 and 6 weeks once the plants reached the second leaf stage.
In the absence of vernalization, winter plants took on average 170 days to head and produced 25 leaves on the main tiller (Fig 6A and 6B). Under this condition, winter MIM172 and vrn1-null headed similarly as winter plants, whereas winter UBIpro:miR172 lines headed after 120 days (50 days earlier) and produced 18 leaves (Fig 6A and 6B). Days to heading and leaf number showed similar profiles across genotypes suggesting that the differences in heading time were due to developmental differences in the timing of the floral transition between the vegetative and reproductive phase and not simply due to differences in the elongation of the stem after the transition. After 6 weeks of vernalization, which is a saturating vernalization treatment for winter Kronos (S8A and S8B Fig), winter UBIpro:miR172, winter MIM172, and winter Kronos plants headed rapidly (33 to 39 days) and showed similar leaf numbers (9 to 9.5 leaves), with only small differences between lines (Fig 6A and 6B). Under the same 6 weeks of cold treatment, vrn1-null plants showed a weaker response to vernalization, headed after 136 days, and produced 24.3 leaves, consistent with previous findings [41].
Fig 6. miR172 levels modulate the flowering response in winter wheat plants.
(A-B) Box plots showing days to heading (A) or number of leaves in the main tiller (B) for vrn1-null, winter Kronos, winter MIM172, and winter UBIpro:miR172 plants grown under LD conditions without vernalization (NV), with 2 (2WV), 3 (3WV) and 6 (6WV) weeks of vernalization (n ≥ 6). (C-H) Expression levels of miR172 (C), AP2L1 (D), AP2L5 (E), VRN1 (F), FT1 (G) and VRN2 (H) determined by qRT-PCR in the 9th leaf of vrn1-null, winter Kronos, winter MIM172, and winter UBIpro:miR172 plants grown under LD conditions in the absence of a vernalization treatment, and after 3 (3WV) and 6 (6WV) weeks of vernalization. After vernalization, the plants were moved to room temperature for 25 days before the fully expanded 9th leaf was collected. SnoR was used as internal reference for miR172, and ACTIN was used as internal reference for the other genes. Data correspond to four independent biological replicates. Different letters above the box-plots indicate significant differences (P <0.05) in Tukey tests performed within each vernalization treatment, except for panel (B) 2WV where a non-parametric Kruskal-Wallis test was used (Data F in S1 Data).
A two-week sub-saturating vernalization treatment accelerated the reproductive transition, with winter Kronos heading after 125 days producing 21.7 leaves and winter UBIpro:miR172 lines heading after 81.5 days producing 15.3 leaves (Fig 6A and 6B). The two-week vernalization treatment produced a stronger acceleration of heading time and reduction in leaf number in winter Kronos (-39.5 days and -3.83 leaves) and UBIpro:miR172 (-37.3 days and -3.3 leaves) than in MIM172 (-19.7 days and -1.2 leaves). Similar trends were observed when plants were exposed to a three-week sub-saturating vernalization treatment (Fig 6A and 6B). Interestingly, the MIM172 winter plants, which showed a weaker response, flowered at a similar time as vrn1-null plants under both sub-saturating vernalization treatments. The flowering differences were reflected in a highly significant genotype x vernalization interaction in factorial ANOVAs for both traits (P < 0.001, Data F in S1 Data). Even when we removed the vrn1-null genotype from the analysis, this interaction remained highly significant (P < 0.001 Data F in S1 Data). This result confirmed that miR172 modulates the flowering response under sub-saturating vernalization treatments in winter wheat.
To further characterize the interaction between vernalization and miR172, we collected the ninth leaf 25 days after the plants were taken out of the cold and performed gene expression analyses. Comparison of miR172 levels in the leaves of winter Kronos and vrn1-null plants vernalized for 6 weeks showed induced expression of miR172 only in the plants with the functional VRN1 allele (Fig 6C), consistent with the previous result after 8 weeks of vernalization (Fig 5J). However, the partial vernalization treatment of 3 weeks was not enough to induce miR172 levels, which showed similar levels in leaves of both winter Kronos and vrn1-null plants (Fig 6C).
The ectopic expression of UBIpro:miR172 in winter plants resulted in higher levels of miR172 in all three vernalization treatments, but were the highest after 6 weeks of vernalization (Fig 6C). MIM172 lines showed lower levels of miR172 expression even after 6 weeks of vernalization. Under these conditions, the changes in miR172 expression were inversely correlated with changes in AP2L expression. Elevated miR172 levels resulted in low AP2L1 levels and reduced miR172 levels in MIM172 plants resulted in elevated levels of AP2L1 and AP2L5 (Fig 6D and 6E). Note that the expression levels of AP2L1 in the NV and 3WV leaves were more than an order of magnitude higher than those of AP2L5.
We also quantified the expression of the flowering genes VRN1, FT1, and VRN2 in the same samples (Fig 6F–6H). Both VRN1 and FT1 showed a gradual increase in expression that correlated with the duration of the vernalization treatment, with the highest values observed after 6 weeks of cold (Fig 6F and 6G). Interestingly, after 3 weeks of vernalization, winter UBIpro:miR172 plants expressed significantly higher levels of FT1 and VRN1, while winter MIM172 expressed lower levels for both genes. After 6 weeks of vernalization, winter Kronos, winter UBIpro:miR172 and winter MIM172 plants expressed similar levels of VRN1, whereas FT1 was still significantly higher in UBIpro:miR172 and not induced in vrn1-null plants. An opposite trend was observed for VRN2, where expression levels decreased as the time in the cold increased, except in vrn1-null plants, which showed constitutive VRN2 expression (Fig 6H). As expected, higher VRN1 transcript levels in winter UBIpro:miR172 plants after 3 weeks of vernalization were associated with lower VRN2 mRNA levels than in winter Kronos. However, after 6 weeks of vernalization, VRN2 was repressed to similar levels in winter Kronos, winter UBIpro:miR172, and winter MIM172 (Fig 6H).
AP2L1 and AP2L5 play significant roles in the modulation of flowering time in winter wheat exposed to sub-saturating vernalization treatments
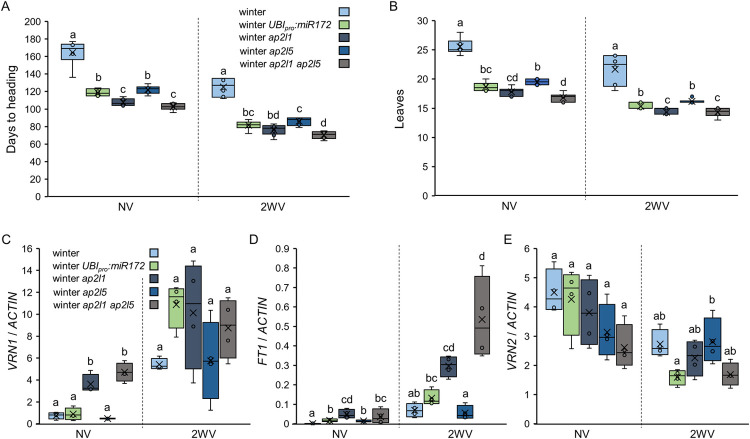
To test if the effects of miR172 on the flowering response to vernalization in winter wheat were mediated by its repression of the AP2L genes, we crossed ap2l5 and ap2l1 mutations with winter Kronos lines and assessed heading time without vernalization or with a sub-saturating 2-week vernalization treatment (Fig 7). Under both conditions, both single ap2l5 and ap2l1 mutants headed 39 to 57 days earlier and had 5.5 to 7.7 fewer leaves than the winter plants and were similar to winter UBIpro:miR172 transgenic plants (Fig 7A and 7B). The ap2l1 mutant was slightly earlier (10–14 days) and had less leaves (1.7–1.8 leaves) than the ap2l5 mutant (Fig 7A and 7B).
Fig 7. AP2L1 and AP2L5 genes modulate the flowering response in winter plants.
(A-E) Winter Kronos, winter UBIpro:miR172, winter ap2l1, winter ap2l5 and winter ap2l1 ap2l5 grown under LD conditions without vernalization (NV) and with 2 (2WV) weeks of vernalization. Box plots showing (A) days to heading or (B) number of leaves in the main tiller (n ≥ 6). (C-E) Expression levels of VRN1 (C), FT1 (D) and VRN2 (E) determined by qRT-PCR in the 9th leaves. After vernalization, the plants were moved to room temperature for 25 days before the fully expanded 9th leaf was collected. ACTIN was used as internal reference. Data correspond to four independent biological replicates. Different letters above the box plots indicate significant differences (P < 0.05) in Tukey tests, except for panels (B) 2WV and (D) NV where non-parametric Kruskal-Wallis tests were used (Data G in S1 Data).
The winter ap2l1 ap2l5 combined mutant showed the earliest heading and fewest number of leaves relative to all other genotypes in the absence of vernalization (102 days and 16.8 leaves) or when partially vernalized (70 days and 14.3 leaves), and in both cases it headed earlier and had fewer leaves than winter UBIpro:miR172 plants (Fig 7A and 7B). It is worth noting that the non-vernalized ap2l1 ap2l5 mutant still headed two months later than fully vernalized winter Kronos plants or non-vernalized spring Kronos, indicating that the ap2l1 ap2l5 mutant reduced but did not abolish the vernalization requirement in wheat.
Consistent with their faster flowering, the non-vernalized ap2l1, and ap2l1 ap2l5 mutant plants showed significantly higher levels of VRN1 and FT1 than the winter control. These differences were maintained after two weeks of vernalization but were significant only for FT1 (Fig 7C and 7D). VRN2 expression was reduced in the mutants, but the differences were not significant (Fig 7E). We did not observe elevated levels of VRN1 and FT1 or a reduction in VRN2 levels in the ap2l5 mutant plants in spite of their earlier heading and reduced leaf number relative to the winter control. However, since the ap2l5 headed slightly later than the other two mutants (Fig 7A and 7B), we cannot rule out similar changes in expression after our sampling point.
Finally, we combined the MIM172 and UBIpro:AP2L-B1 transgenes in the winter Kronos background. We expected both MIM172 and UBIpro:AP2L-B1 to delay flowering in response to vernalization treatments; thus, we analyzed the progeny under a 4-week partial vernalization experiment, which is closer to a saturating 6-week vernalization treatment. Heading of the MIM172 plants was delayed relative to the winter Kronos line, but both MIM172 and winter Kronos were earlier than the vrn1-null plants (S10 Fig and Data O in S1 Data). Interestingly, winter plants harboring both MIM172 and UBIpro:AP2L-B1 were later than MIM172 plants and headed at a similar time as the vrn1-null plants (S10 Fig).
Taken together, these results show that in winter wheat cultivars, the levels of miR172 and AP2L genes affect the duration of cold exposure required to induce changes in the expression of VRN1, VRN2, and FT1 genes, which in turn can modulate the flowering response to sub-saturating vernalization conditions.
Discussion
AP2L genes function as flowering repressors
In this work, we used a combination of genetic, transgenic, and expression analyses to demonstrate that the miR172-AP2L module plays important roles in the regulation of flowering time in wheat through interaction with central players of the vernalization pathway.
AP2L proteins are transcription factors that function as repressors of the flowering transition in several species, including eudicot species such as Arabidopsis and monocots such as rice [54]. In these species, the AP2L gene family includes several members that usually have overlapping roles in the regulation of the flowering transition. Arabidopsis for example contains six AP2L genes (TOE1, TOE2, TOE3, SCHLAFMUTZE (SMZ), SCHNARCHZAPFEN (SNZ), and AP2), and a previous study showed that the toe1 toe2 double mutants flowered earlier than either single mutant, but still it was not as rapid flowering as the 35Spro:miR172 overexpressing lines [58]. A more recent study showed that the toe1 toe2 smz snz toe3-1 ap2-12 sextuple mutant phenocopied the 35Spro:miR172 plants very closely [55].
Our work also showed overlapping roles for AP2L1 and AP2L5 genes in the control of the flowering transition in wheat, with both single and combined ap2l1 ap2l5 mutants flowering earlier than the sister wild type lines in spring and winter wheat. The ap2l1 ap2l5 mutant was earlier than the single mutants and the UBIpro:miR172 transgenic plants. The relatively mild effect of UBIpro:miR172 on heading time can be an indirect effect of our selection of a fertile transgenic line with a weak floret phenotype, which likely expresses intermediate levels of the transgene. In our previous study, strong overexpression of miR172 in Kronos resulted in floret defects and sterile plants that could not be propagated [84].
In addition to its effect on flowering time, AP2L5 affected inflorescence and flower development, and the ap2l5 mutant defects were magnified when combined with ap2l2 mutants [84]. However, ap2l1 mutants developed normal inflorescences (S2 Fig). Similar observations were described in Arabidopsis, with TOE1 (ortholog of AP2L1) and TOE2 affecting flowering time, and AP2 (closer to AP2L2 [84]) regulating both flowering time and flower development [55,58,72]. These results suggest an ancient sub-functionalization of these two AP2L clades that is still maintained in Arabidopsis and wheat.
The mechanisms by which AP2L transcription factors control flowering are best described in Arabidopsis. Chromatin immunoprecipitation analyses revealed that TOE1 and SMZ proteins can bind to FT promoter sequences and inhibit its expression in leaves [56,57]. In addition, these genome-wide experiments showed that SMZ and AP2 repress many other flowering time regulators acting downstream of FT in the leaves and the SAM [55,56]. Besides this direct transcriptional regulation of FT and other flowering genes, TOE proteins were shown to inhibit CO activity in Arabidopsis [57], as they can interact with the transcriptional activation domain of CO and affect CO protein stability. Similarly, our results indicate that miR172 can induce FT1 expression in the leaves, likely through the repression of AP2L genes, and that this mechanism is independent of VRN1 and VRN2, or changes in the expression of PPD1. However, we currently do not know if this transcriptional regulation of FT1 results from an indirect effect or is mediated by direct binding of AP2L proteins to FT1 regulatory sequences.
Variable contributions of AP2L genes to flowering in spring and winter wheats under different photoperiods
VRN2 and AP2L genes are repressors of flowering that likely act by repressing FT1 transcription in the leaves. However, the elimination of both types of FT1-repressing genes in UBIpro:miR172 vrn2 vrn1 plants did not accelerate flowering to the same extent observed in backgrounds harboring the spring Vrn1 allele [UBIpro:miR172 vrn2 vrn1: ~55 days (Fig 2K) vs. UBIpro:miR172 vrn2 Vrn1: ~35 days (Fig 2F)]. This result indicates that VRN1 can accelerate flowering independently of VRN2 and AP2L genes. This could be mediated by a direct regulation of FT1 in the leaves, a hypothesis supported by VRN1 promotion of FT1 expression in leaves of vrn2-null plants [42], and by the binding of VRN1 to the FT1 promoter in a CHIP-seq experiment in barley [86]. In addition, VRN1 was shown to promote the transition of the SAM between the vegetative and early reproductive stages even in the absence of FT1¸ but FT1 expression was still necessary at later stages for normal spike development and stem elongation [91,92].
The relative contributions of VRN2 and AP2L genes to the repression of flowering in wheat depend on the genetic background. In spring cultivars, both pathways contribute similarly to the repression of flowering, and UBIpro:miR172 vrn2 plants showed an additive effect with very rapid flowering (Fig 2). However, in winter cultivars grown in the absence of vernalization, VRN2 plays a more predominant role, since vrn2 vrn1 plants flowered within 65 days, while UBIpro:miR172 vrn1 required 120 days to flower. The prominent role of VRN2 is also supported by the observation that, in both diploid wheat and barley, major QTLs for spring growth habit have been mapped to multiple loss-of function alleles of VRN2 [29,35]. Our results indicate that although miR172 and AP2L genes play a more limited role than VRN2 in the repression of flowering in non-vernalized winter plants, they can modulate the flowering response under non-saturating vernalization conditions.
Interactions between AP2L genes and vernalization have also been described in the perennial brassica species A. alpina. In this species, the orthologs of Arabidopsis AP2 [60,61] and TOE2 [93] regulate the age-dependent response to vernalization and contribute to the perennial growth habit. However, whereas loss-of-function mutations in A. alpina ortholog of AP2 were sufficient to confer a spring growth habit [60,61], winter wheat plant overexpressing UBIpro:miR172 or carrying ap2l1 ap2l5 mutations showed a reduced but still strong vernalization requirement (Figs 6A, 6B, 7A, and 7B). These observations confirm a conserved role of AP2L genes as flowering repressors and indicate that their relative contribution to the vernalization requirement can vary among species.
Our results also show that the miR172-AP2L module regulates Kronos flowering time under SD, and that the effects were stronger than under LD (Figs 1 and S1). In maize, where some varieties behave as SD plants, a major flowering-time QTL, named Vegetative to generative transition 1 (Vgt1), was mapped to a 2-kb conserved noncoding region positioned 70 kb upstream of ZmTOE1 [63]. Vgt1 acts as a cis-acting regulatory element that affects the transcript expression levels of ZmTOE1 and flowering time in maize. Interestingly, a recently identified short-day flowering time response QTL on wheat chromosome 1BS has also been linked to AP2L-B1 (TaTOE1) [94]. Since the AP2L-B1 mutant alleles were associated with earlier flowering, the authors suggested that AP2L-B1 likely works as a flowering repressor, which agrees with the results presented here.
These natural AP2L1 alleles, together with the mutant alleles generated in this work, provide wheat breeders with additional tools to fine-tune wheat flowering time to specific environments.
Vernalization and plant age control miR172 and AP2L expression in leaves
miR156-SPLs and miR172-AP2Ls are two conserved miRNA modules in plants that display a complementary expression pattern in the shoot with plant age [78,82,83]. Our results showed similar complementary expression patterns for miR156 and miR172 in spring wheat. In MIM156 plants with reduced miR156 activity, miR172 expression is higher in juvenile leaves (Fig 4), while adult leaves of plants overexpressing miR156 have lower levels of miR172 and higher levels of AP2L1 (Fig 3). These results indicate that miR156 is necessary for the correct expression pattern of miR172 and AP2L1. However, these interactions are altered in winter wheat backgrounds. While the expression profile of miR156 was similar in spring and winter cultivars, the expression of miR172 differed between these two backgrounds. In spring plants, miR172 expression increased rapidly reaching high levels at early stages following the flowering transition, whereas this increase with age was delayed in winter wheat compared with spring wheat. These results suggest that miR156 and miR172-AP2Ls modules are connected, but also that other mechanisms may regulate the expression profile of miR172 downstream of miR156.
We speculate that during the induction of the flowering transition several feedback loops act to reinforce changes in gene expression and to produce a robust shift in the developmental program. Vernalizing winter plants for several weeks promotes rapid induction of miR172 and downregulation of AP2L1 similar to that observed in spring cultivars. Interestingly, similar results were observed in the perennial brassica A. alpina. In this species the accumulation of miR156 is reduced in the shoot apex of older plants that acquire competence to flower in long days, but miR172 expression is low. Vernalization is required to induce flowering and to promote an increase in miR172 levels in the shoot apex [61].
Another interesting observation from this work was that in winter plants AP2L1 expression was not downregulated with plant age even when miR172 expression increased. It has been shown that miR172 levels can reduce AP2L protein levels without affecting transcript levels [58,72]. It was suggested that miR172 might repress the translation of AP2L mRNA [58,72], or that feedback loops involving AP2L genes controlling their own transcription act to keep transcript levels constant [85]. However, downregulation of AP2L1 expression by vernalization (Figs 5 and 6) or in the flag leaf of late flowering winter plants grown without vernalization (Fig 5) suggest that other mechanisms may also exist in wheat to repress AP2L gene expression.
A recent study in Arabidopsis has shown that FUL directly represses the transcription of several AP2L genes such as SNZ, TOE1, and AP2 during inflorescence development in parallel to miR172 [90]. An additional Arabidopsis study indicated that FUL and miR172 control AP2L gene activity at both transcriptional and post-transcriptional levels to promote the floral transition [70], which could provide a rapid and robust mechanism to deplete AP2L genes as flowering proceeds. This seems to be also the case in wheat, where vernalization of winter wheat lines results in the downregulation of AP2L1, but only in lines with active VRN1 genes. The high levels of AP2L genes in vernalized vrn1-null plants relative to the vernalized plants with a functional VRN1 allele indicates that VRN1 plays a critical role in the transcriptional downregulation of AP2L1. Since the overexpression of miR172 had no significant effects on the transcriptional regulation of AP2L1 and AP2L5 (S3 Fig), we think that the upregulation of miR172 by VRN1 (Fig 5J) is insufficient to explain the transcriptional downregulation of AP2L1 by VRN1 (Fig 5K).
In winter wheat, vernalization results in the induction of the expression of VRN1 and its closest paralogs FUL2 and FUL3 (Figs 6 and S9G–S9I). These three genes are also induced in older winter plants, even in the absence of vernalization (Figs 5, S9D, and S9E), and the timing of their inductions coincides with the downregulation of AP2L1 and AP2L5 (Figs 5F and S9C), providing additional support for the role of VRN1 in the direct or indirect downregulation of AP2L genes in the leaves.
We speculate that the downregulation of AP2Ls at later developmental stages could reflect an additional role of these genes on internode elongation. Transgenic wheat plants expressing MIM172 have higher AP2Ls expression and shorter internodes than the non-transgenic controls [66]. Similarly, a dominant mutation in the miR172 target site of AP2L2 in barley negatively regulates peduncle elongation during the reproductive transition [95]. Since the downregulation of AP2Ls in flag leaves was associated with an increase in FT1 expression (Fig 5), and FT1 is required to promote internode elongation in wheat [92], we speculate that AP2Ls may control internode elongation by controlling FT1 expression in the leaves.
miR156 regulates flowering by controlling miR172 and other genes
In Arabidopsis, miR156 negatively regulates the expression of multiple SPL genes, which directly promote the induction of miR172 expression and the initiation of the reproductive phase [65,78,96–98]. In addition, SPL transcription factors can also modulate flowering by miR172-independent pathways. For example, Arabidopsis SPL3, SPL9 and SPL15 promote the expression of SOC1, FUL, AP1 and LFY in the SAM [96,98,99], and these interactions are relevant to flowering promotion under non-inductive SD conditions.
In wheat, overexpression of miR156 also results in the repression of several SPL genes [88], suggesting that the miR156-SPL module is conserved in wheat. The significantly earlier flowering observed in the transgenic wheat plants including both MIM156 and MIM172 relative to those including only MIM172 (S7 Fig) indicates that miR156 can also regulate flowering time by miR172-independent pathways. Our data indicate that, in addition to miR172, FUL2 and FUL3 might participate in the flowering responses downstream of the miR156-SPL module. In adult leaves of the late flowering plants overexpressing miR156, FUL2 and FUL3 expression is reduced (Fig 3H and 3I), similar to miR172. Moreover, in the leaves of non-vernalized winter plants, FUL2 and FUL3 expression is induced a couple of weeks earlier (W12, S9D and S9E Fig) than FT1 and VRN-B1 (W14, Fig 5B and 5C).
Overexpression of FUL2 using the maize UBIQUITIN promoter in Kronos resulted in accelerated flowering [100]; thus, it would be interesting to test whether SPL transcription factors promote flowering in non-vernalized winter plants through the induction of FUL2 and FUL3 expression.
Integrating the miR172-AP2L regulatory module into the wheat flowering network
Flowering time must occur at an optimal time to maximize reproductive success. This precision is achieved through a complex regulatory network that senses, translates and integrates different signals into the regulation of a few central flowering genes. In wheat, flowering is promoted by long days and vernalization and both pathways converge on the activation of FT1 in the leaves (Fig 8). We incorporate into this model the conserved miR156-miR172-AP2L module, which constitutes another pathway to regulate FT1 expression in the leaves (Fig 8).
Fig 8. Working model for flowering regulation in wheat.
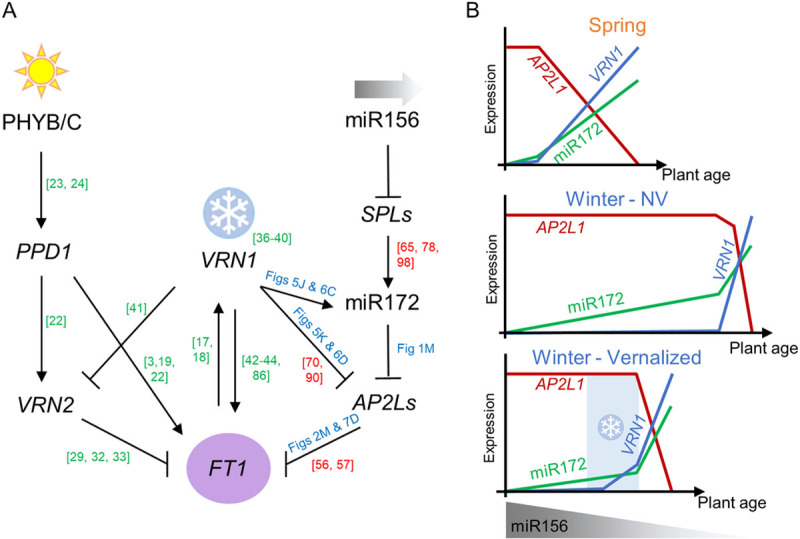
(A) Flowering network integrating photoperiod, vernalization and age signals into the regulation of FT1 expression in wheat leaves. Arrows indicate promotion of gene expression and lines ending in a crossed-bar repression. Numbers in brackets indicate the references supporting the different interactions (red, indicates Arabidopsis references). The critical figures supporting the interactions presented in this paper are indicated in blue. (B) Model illustrating the expression profile of miR172 and AP2L1 with plant development. Top panel shows expression profiles of miR172 and AP2L1 in spring wheat cultivars, where VRN1 is expressed in the absence of cold. The middle panel shows expression profiles of miR172 and AP2L1 in a winter wheat background without vernalization (Winter—NV). The bottom panel shows expression profiles in vernalized winter plants (Winter–Vernalized). In this background, VRN1 expression after vernalization (light blue region) promotes miR172 upregulation and AP2L1 downregulation. In both spring and winter cultivars, miR156 expression in leaves is downregulated with plant age.
In this model, both VRN2 and AP2Ls repress FT1 in the leaves to prevent flowering in the fall. In spring cultivars, the levels of AP2L genes are down-regulated developmentally by the sequential action of miR156 and miR172, which promotes a rapid flowering transition in spring cultivars. However, in winter wheat plants, the upregulation of miR172 and the downregulation of AP2L genes requires the induction of VRN1 during vernalization, which results in an additional regulatory layer that delays the upregulation of FT1 and prevents premature flowering in the fall. In turn, changes in the dosage or repression activity of AP2Ls affect flowering time in winter wheat backgrounds. Under sub-saturating conditions, plants with lower dosage or activity of AP2L require less VRN1 and lower reductions in VRN2 expression (less cold) to induce FT1 expression.
Taken together, our results show that conserved flowering genes and temperate grass-specific genes are highly interconnected in a network that integrates plant age, vernalization and photoperiod to mediate a timely flowering response (Fig 8). Our results also show that modulation of the AP2L dosage or activity may be a valuable tool to modulate the flowering response of winter cultivars as winters become milder due to climate change.
Materials and methods
Plant materials and growth conditions
The tetraploid wheat variety Kronos used in this study has a spring growth habit determined by the Vrn-A1c allele (intron 1 deletion), and a reduced photoperiod response conferred by the Ppd-A1a allele (promoter deletion). Kronos also has the AP2L-5A Q allele, which confers the subcompact spike phenotype and the free-threshing character, and a non-functional AP2L-5B gene [66]. TILLING populations of Kronos and Cadenza (hexaploid) mutagenized with ethyl methanesulfonate (EMS) [101] were used to screen for mutants in AP2L1. The two selected truncation mutations in AP2L1 homeologs, and the mutation in the miR172 target site in AP2L-A1 were confirmed in M4 grains using genome specific primers described in S3 Table.
The mutant M4 plants were crossed at least two times with the parental wild type Kronos to reduce the mutation background before phenotypic analysis. The mutation CAD161 was identified in the hexaploid wheat Cadenza. Since Kronos x Cadenza crosses result in hybrid necrosis, we used a bridge cross between CAD161 mutant to an F2 plant from the cross between the hexaploid line Insignia and the tetraploid Kronos. We then backcrossed the F1 to Kronos twice.
The vrn-a1 and vrn1-null lines [41], and ap2l5 mutant line K3946 [66] were described before. For all experiments, grains were first cold imbibed for 2–4 days at 4°C before transferring them to room temperature. The plants were grown in one-gallon pots in PGR15 growth chambers (Conviron) adjusted to 16 h of light (22°C) and 8 h of darkness (18°C) (Long Day condition or LD) or 8 h of light (22°C) and 16 h of darkness (18°C) (Short Day condition or SD). Intensity of the sodium halide lights measured at plant height was (~260–300 μM m-2 s-1).
Plants at the second leaf stage were vernalized in one-gallon pots in a cold chamber (Conviron) with temperatures averaging 5°C and daylength set to 16 h of light / 8 h of darkness. Intensity of the light during the light phase at plant height was 230 μM m-2 s-1. Vernalization treatments of different length were used in different experiments: In Figs 5G–5K and S9F–S9I, plants were vernalized for 8 weeks; in Fig 6A and 6B, plants were vernalized for 2, 3 and 6 weeks; in Fig 6C–6H, plants were vernalized for 3 and 6 weeks; in S10 Fig, plants were vernalized for 2 weeks; and in S11 Fig, plants were vernalized for 4 weeks.
qRT-PCR
In all experiments we collected the last fully expanded leaf in the main tiller, and harvested samples 4 hours after lights were turned on, except in Fig 6 where leaves were harvested in the middle of the photoperiod (ZT8). RNA samples were extracted using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). We followed Protocol A that allows purification of total RNA including small RNA molecules. Total RNA was treated with RQ1 RNase-free DNase (Promega). cDNA synthesis was carried out using SuperScript II Reverse Transcriptase (Invitrogen). mRNAs were reverse transcribed starting from 1 μg of total RNA and using OligodTv primer. Mature miR172 and miR156 levels were determined by stem-loop qRT-PCR as described previously [102], specific stem-loop oligos to mature miR156 and miR172 are described in S3 Table. The reverse primer for the small nucleolar RNA 101 (snoR101), which is the reference used to normalize miRNA in qRT-PCR, was also included in the reverse transcription. RNA extraction and cDNA synthesis for the vernalization time course experiments in Figs 6 and 7 were done as described in [44]. The product from the 1st strand synthesis was diluted 1 in 20, and 5 μl of diluted cDNAs was used in qRT-PCR reaction. Quantitative PCR was performed using SYBR Green and a 7500 Fast Real-Time PCR system (Applied Biosystems). The ACTIN gene was used as an endogenous control for mRNAs, and SnoR101 for miRNAs. Primers for the different genes tested are listed in S3 Table.
Vectors
The coding region of the AP2L-B1 gene was amplified from Kronos cDNA using primers in S3 Table and cloned into the pENTR vector (Invitrogen). It was then subcloned into the pLC41 vector downstream of the maize UBIQUITIN promoter (UBIpro) with a C-terminal HA tag (henceforth UBIpro:AP2L-B1). To generate miR156 overexpressing lines, the miR156b,c locus (B genome) was amplified by PCR from Kronos genomic DNA using primers in S3 Table. The PCR product was cloned into pDONR (Invitrogen) and then sub-cloned into pLC41 downstream of UBIpro. Since UBIpro:miR156b,c plants were sterile, a LhG4/pOp two component system was developed in wheat to maintain a stable miR156 overexpressing line. LhG4 is a synthetic transcription factor that binds to the synthetic pOp promoter and activates transcription [89]. The LhG4 coding sequence was cloned into pLC41 vector downstream of the maize UBIpro to generate the driver vector (UBIpro:LhG4). The UBIpro in UBIpro:miR156b,c vector was replaced by pOp to generate the responder vector (pOp:miR156). Finally, UBIpro:LhG4 and pOp:miR156 plants were crossed, and the phenotypic and molecular analysis were performed in the F1 plants.
To generate the artificial MIM156, the wheat ortholog of the natural miR399-target-mimic IPS1 gene described in Arabidopsis [103] was synthesized and the sequence complementary to miR399 was replaced by a sequence complementary to miR156. The complete sequence of MIM156 is provided in S11 Fig. The MIM156 construct was cloned into the binary vector pLC41 downstream of the maize UBIpro. Lines MIM172 and UBIpro:miR172 were described before [66].
Wheat transformation
Transgenic wheat plants were generated at the UC Davis Plant Transformation Facility (http://ucdptf.ucdavis.edu/) using the Japan Tobacco (JT) technology licensed to UC Davis. Immature embryos from Kronos were transformed using Agrobacterium EHA105. Selection of transgenic plants was conducted using hygromycin, and transgene insertion was validated by DNA extraction and PCR.
Statistical analyses
The raw data and descriptive statistics for all graphs and tables presented in this study are provided in the S1 Data file. Mean comparison were done using Tukey tests that compare all means with each other. In experiments including a wild-type (Wt) control Dunnett tests were also performed and are included in the S1 Data to provide a more precise estimate of the significance of the differences with the Wt. In all statistical comparisons, homogeneity of variance using Levene’s test and normality of residuals using the Shapiro-Wilk test were first tested. If the data did not fit the assumptions of the analysis of variance (ANOVA), power transformations were used to restore them. When power transformations could not satisfy both assumptions, non-parametric Kruskal-Wallis tests were used. All statistical analyses were performed using SAS version 9.4.
Distribution of the data presented with box-plots including individual data points were generated with Excel. The middle line of the box represents the median and the x represents the mean. The bottom line of the box represents the first quartile and the top line the third quartile. The whiskers extend from the ends of the box to the minimum value and maximum value.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(A) Nine-week-old UBIpro:miR172, wild type Kronos (Wt) and MIM172 plants grown under SD. Scale bar = 10 cm. (B-C) Box plots showing days to heading (B; n ≥ 7) and the number of leaves produced by the main tiller (C; n ≥ 9) in UBIpro:miR172, wild type Kronos (Wt) and MIM172 plants grown under SD. (D-G) Box plots showing expression levels of VRN1 (D), FT1 (E), VRN2 (F), and PPD1 (G) determined by qRT-PCR in the 3rd (L3) and 6th (L6) leaves of UBIpro:miR172, wild type Kronos (Wt) and MIM172 plants growing under SD. ACTIN was used as an internal reference. Data correspond to four independent biological replicates. Different letters above the box plots indicate significant differences based on Tukey tests (P < 0.05). Raw data and statistical tests in Data H in S1 Data.
(TIF)
(A) Main spikes and (B) central spikelets from wild type Kronos (Wt), ap2l1, ap2l5 and ap2l1 ap2l5 plants. Red asterisks in ap2l5 and ap2l1 ap2l5 spikelets indicate first empty lemmas.
(TIF)
(A-B) Box plots showing AP2L1 (A) and AP2L5 (B) expression levels in the 5th leaf of UBIpro:miR172, wild type Kronos (Wt) and MIM172 plants grown under LD (same biological samples used to quantify miR172 expression in Fig 1B). Transcript levels were determined by qRT-PCR using ACTIN as internal reference. Data correspond to four independent biological replicates. Different letters above the box plots indicate significant differences based on Tukey tests (P < 0.05). Raw data and statistical tests in Data I in S1 Data.
(TIF)
(A-B) Expression levels of FUL2 (A) and FUL3 (B) determined by qRT-PCR in the 1st (L1), 3rd (L3), 5th (L5) and 7th (L7) leaves of UBIpro:miR172, wild type Kronos (Wt) and MIM172 plants grown under LD. ACTIN was used as internal reference. Data correspond to four independent biological replicates. Different letters above the data point for each leaf indicate significant differences based on Tukey tests (P < 0.05). The color of the letter corresponds to the color of the genotype. Raw data and statistical tests in Data J in S1 Data.
(TIF)
(A-C) Box plots showing the expression levels of VRN1 (A), FT1 (B) and VRN2 (C) determined by qRT-PCR in the 5th leaf of F2 plants segregating for UBIpro:AP2L-B1 and MIM172 transgenes grown under LD. ACTIN was used as internal reference. Data correspond to four independent biological replicates. Different letters above the box plots indicate significant differences based on Tukey tests (P < 0.05). Raw data and statistical tests in Data K in S1 Data.
(TIF)
(A) Scheme showing the cassette including the maize UBIQUITIN promoter (UBIpro) and the sequences corresponding to the miR156b,c (B genome) locus. The predicted secondary structure for the miR156b,c sequence is shown below. Note the three stem-loop structures, including the three miR156/miR156* duplexes, corresponding to three miR156 precursors in tandem. (B) Wild type Kronos plant (Wt, left) and four independent transgenic T0 lines expressing UBIpro:miR156b,c grown under LD. Scale bar = 10 cm.
(TIF)
(A-B) Box plots showing days to heading (A; n ≥ 9) and the number of leaves produced by the main tiller before the transition to the spike (B; n ≥ 10) for wild type Kronos (Wt), MIM156, MIM172 and MIM156 MIM172 plants growing under LD conditions. Different letters above the box plots indicate significant differences (P ≤ 0.05) in pair-wise Kruskal-Wallis tests. Raw data and statistical tests in Data L in S1 Data.
(TIF)
(A-B) Box plots showing the days to heading (A) and the number of leaves produced by the main tiller before the transition to the spike (B) for winter Kronos (vrn-A1) plants grown under LD conditions without vernalization (NV), with 2 (2WV), 4 (4WV), 6 (6WV) and 8 (8WV) weeks of vernalization. Different letters over the box plots indicate significant differences in Kruskal-Wallis non-parametric pairwise tests at P < 0.05. Raw data and statistical tests in Data M in S1 Data.
(TIF)
(A) Six-week-old spring and winter Kronos plants growing under LD conditions without vernalization. Scale bar = 10 cm. (B) Box plots showing the days to heading for spring and winter Kronos plants grown under LD conditions without vernalization (t-test, n ≥ 6). (C-E) Time course of AP2L5 (C), FUL2 (D) and FUL3 (E) expression levels determined by qRT-PCR in spring and winter Kronos plants grown under LD conditions in the absence of vernalization treatment. W1 = week1, 1st leaf; W5 = week 5, 7th leaf; W10 = week 10, leaves 10th-11th; W12 = week 12, leaves 13th-14th; W14 = week 14, leaves 15th-16th; W16 = week 16, leaves 18th-19th; W18, week 18, Flag leaves (20th-23rd). ACTIN was used as internal reference. Data correspond to four independent biological replicates. * = P ≤ 0.05 in t-test comparing expression at the same leaf in spring vs winter plants, except for panel (E) where a non-parametric Kruskal-Wallis test was used. (F-I) Box plots showing the expression of AP2L5 (F), VRN-B1 (G), FUL2 (H) and FUL3 (I) determined by qRT-PCR in the 7th (L7) leaf of winter Kronos without vernalization, and the 7th leaf of winter Kronos and vrn1-null mutants vernalized for 8 weeks and then moved to room temperature for two weeks. Samples from the 7th leaf were collected when the leaves were fully expanded. ACTIN was used as internal reference. Data correspond to at least 5 independent biological replicates. (F) Same letters indicate lack of significant differences in Tukey tests. (H) No expression of FUL2 was detected for the non-vernalized winter Kronos and the vrn1-null samples so no statistical test was performed. (G and I) No expression of VRN-B1 and FUL3 was detected for the non-vernalized winter Kronos sample, so a t-test was performed for the other two samples (*** = P < 0.001). Raw data and statistical tests in Data N in S1 Data.
(TIF)
(A-B) Box plots showing days to heading (A) and number of leaves produced by the main tiller (B) in plants grown under LD conditions with 4 weeks of vernalization. vrn1-null = no functional VRN1 genes, winter Kronos, winter line with MIM172 transgene, winter line with MIM172 and UBIpro:AP2L-B1 transgenes. Different letters above the box plots indicate significant differences (P < 0.05) in Tukey tests. Raw data and statistical tests in Data O in S1 Data.
(TIF)
The sequence complementary to miR156 is in red.
(TIF)
Data A in S1 Data. Supporting data for Fig 1. Data B in S1 Data. Supporting data for Fig 2. Data C in S1 Data. Supporting data for Fig 3. Data D in S1 Data. Supporting data for Fig 4. Data E in S1 Data. Supporting data for Fig 5. Data F in S1 Data. Supporting data for Fig 6. Data G in S1 Data. Supporting data for Fig 7. Data H in S1 Data. Supporting data for S1 Fig. Data I in S1 Data. Supporting data for S3 Fig. Data J in S1 Data. Supporting data for S4 Fig. Data K in S1 Data. Supporting data for S5 Fig. Data L in S1 Data. Supporting data for S7 Fig. Data M in S1 Data. Supporting data for S8 Fig. Data N in S1 Data. Supporting data for S9 Fig. Data O in S1 Data. Supporting data for S10 Fig.
(XLSX)
Acknowledgments
We thank Mariana Padilla and Xiaoqin Zhang for excellent technical support. We thank Dr. David Jackson for providing the vectors with LhG4 and pOp sequences.
Data Availability
All the raw data supporting every figure and every table is provided as Supplemental Data.
Funding Statement
J.D. acknowledges support from the Howard Hughes Medical Institute (HHMI Researcher Funding, https://www.hhmi.org/) and by competitive Grants 2022-67013-36209 and 2022-68013-36439 (WheatCAP) from the United States Department of Agriculture, National Institute of Food and Agriculture (https://nifa.usda.gov/). J.M.D. was supported by a fellowship (LT000590/2014-L) of the Human Frontier Science Program (hfsp.org). D.P.W is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation (http://www.lsrf.org/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol. 2015;66:441–64. doi: 10.1146/annurev-arplant-043014-115555 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ream TS, Woods DP, Amasino RM. The molecular basis of vernalization in different plant groups. Cold Spring Harb Symp Quant Biol. 2012;77:105–15. doi: 10.1101/sqb.2013.77.014449 . [DOI] [PubMed] [Google Scholar]
- 3.Pearce S, Shaw LM, Lin H, Cotter JD, Li C, Dubcovsky J. Night-break experiments shed light on the Photoperiod1-mediated flowering. Plant Physiol. 2017;174(2):1139–50. doi: 10.1104/pp.17.00361 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouche F, Woods DP, Amasino RM. Winter memory throughout the plant kingdom: different paths to flowering. Plant Physiol. 2017;173(1):27–35. doi: 10.1104/pp.16.01322 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chouard P. Vernalization and its relations to dormancy. Annu Rev Plant Phys. 1960;11: 191–238. doi: 10.1146/annurev.pp.11.060160.001203 PMID:WOS:A1960WQ99100010. [DOI] [Google Scholar]
- 6.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–39. doi: 10.1038/nrg3291 . [DOI] [PubMed] [Google Scholar]
- 7.Cao S, Luo X, Xu D, Tian X, Song J, Xia X, et al. Genetic architecture underlying light and temperature mediated flowering in Arabidopsis, rice, and temperate cereals. New Phytol. 2021;230(5):1731–45. doi: 10.1111/nph.17276 . [DOI] [PubMed] [Google Scholar]
- 8.Kim DH, Doyle MR, Sung S, Amasino RM. Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009;25:277–99. doi: 10.1146/annurev.cellbio.042308.113411 . [DOI] [PubMed] [Google Scholar]
- 9.Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, et al. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci U S A. 2006;103(51):19581–6. doi: 10.1073/pnas.0607142103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nitcher R, Distelfeld A, Tan C, Yan L, Dubcovsky J. Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol Genet Genomics. 2013;288(5–6):261–75. doi: 10.1007/s00438-013-0746-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brassac J, Muqaddasi QH, Plieske J, Ganal MW, Röder MS. Linkage mapping identifies a non-synonymous mutation in FLOWERING LOCUS T (FT-B1) increasing spikelet number per spike. Sci Rep. 2021;11(1):1585. doi: 10.1038/s41598-020-80473-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballerini ES, Kramer EM. In the Light of Evolution: A reevaluation of conservation in the CO-FT regulon and its role in photoperiodic regulation of flowering time. Front Plant Sci. 2011;2:81. doi: 10.3389/fpls.2011.00081 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–3. doi: 10.1126/science.1141752 . [DOI] [PubMed] [Google Scholar]
- 14.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316(5827):1033–6. ISI:000246554000048. doi: 10.1126/science.1141753 [DOI] [PubMed] [Google Scholar]
- 15.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309(5737):1052–6. doi: 10.1126/science.1115983 ISI:160999790100039. [DOI] [PubMed] [Google Scholar]
- 16.Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309(5737):1056–9. ISI:000231230100040. doi: 10.1126/science.1114358 [DOI] [PubMed] [Google Scholar]
- 17.Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008;55(4):543–54. doi: 10.1111/j.1365-313X.2008.03526.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Lin H, Dubcovsky J. Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 2015;84(1):70–82. doi: 10.1111/tpj.12960 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310(5750):1031–4. doi: 10.1126/science.1117619 . [DOI] [PubMed] [Google Scholar]
- 20.Higgins JA, Bailey PC, Laurie DA. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One. 2010;5(4):e10065. doi: 10.1371/journal.pone.0010065 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campoli C, Shtaya M, Davis SJ, von Korff M. Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 2012;12:97. doi: 10.1186/1471-2229-12-97 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw LM, Li C, Woods DP, Alvarez MA, Lin H, Lau MY, et al. Epistatic interactions between PHOTOPERIOD1, CONSTANS1 and CONSTANS2 modulate the photoperiodic response in wheat. PLoS Genet. 2020;16(7):e1008812. doi: 10.1371/journal.pgen.1008812 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen A, Li C, Hu W, Lau MY, Lin H, Rockwell NC, et al. Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc Natl Acad Sci U S A. 2014;111(28):10037–44. doi: 10.1073/pnas.1409795111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woods DP, Ream TS, Minevich G, Hobert O, Amasino RM. PHYTOCHROME C is an essential light receptor for photoperiodic flowering in the temperate grass, Brachypodium distachyon. Genetics. 2014;198(1):397–408. doi: 10.1534/genetics.114.166785 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet. 2007;115(5):721–33. doi: 10.1007/s00122-007-0603-4 . [DOI] [PubMed] [Google Scholar]
- 26.Wilhelm EP, Turner AS, Laurie DA. Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.). Theor Appl Genet. 2009;118(2):285–94. doi: 10.1007/s00122-008-0898-9 . [DOI] [PubMed] [Google Scholar]
- 27.Alqudah AM, Sharma R, Pasam RK, Graner A, Kilian B, Schnurbusch T. Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS One. 2014;9(11):e113120. doi: 10.1371/journal.pone.0113120 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stracke S, Haseneyer G, Veyrieras JB, Geiger HH, Sauer S, Graner A, et al. Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theor Appl Genet. 2009;118(2):259–73. doi: 10.1007/s00122-008-0896-y . [DOI] [PubMed] [Google Scholar]
- 29.Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303(5664):1640–4. doi: 10.1126/science.1094305 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–7. doi: 10.1038/ng.143 . [DOI] [PubMed] [Google Scholar]
- 31.Woods DP, McKeown MA, Dong Y, Preston JC, Amasino RM. Evolution of VRN2/Ghd7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol. 2016;170(4):2124–35. doi: 10.1104/pp.15.01279 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Distelfeld A, Tranquilli G, Li C, Yan L, Dubcovsky J. Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 2009;149(1):245–57. doi: 10.1104/pp.108.129353 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kippes N, Chen A, Zhang X, Lukaszewski AJ, Dubcovsky J. Development and characterization of a spring hexaploid wheat line with no functional VRN2 genes. Theor Appl Genet. 2016;129(7):1417–28. doi: 10.1007/s00122-016-2713-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karsai I, Szucs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, et al. The Vrn-H2 locus is a major determinant of flowering time in a facultative x winter growth habit barley (Hordeum vulgare L.) mapping population. Theor Appl Genet. 2005;110(8):1458–66. doi: 10.1007/s00122-005-1979-7 . [DOI] [PubMed] [Google Scholar]
- 35.von Zitzewitz J, Szucs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, et al. Molecular and structural characterization of barley vernalization genes. Plant Mol Biol. 2005;59(3):449–67. doi: 10.1007/s11103-005-0351-2 . [DOI] [PubMed] [Google Scholar]
- 36.Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of wheat vernalization gene VRN1. Proc Natl Acad Sci USA. 2003;100:6263–8. doi: 10.1073/pnas.0937399100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci U S A. 2003;100(22):13099–104. doi: 10.1073/pnas.1635053100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu D, Szűcs P, Yan L, Helguera M, Skinner J, Hayes P, et al. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4 [DOI] [PubMed] [Google Scholar]
- 39.Kippes N, Guedira M, Lin L, Alvarez MA, Brown-Guedira GL, Dubcovsky J. Single nucleotide polymorphisms in a regulatory site of VRN-A1 first intron are associated with differences in vernalization requirement in winter wheat. Mol Genet Genomics. 2018;293:1231–43. doi: 10.1007/s00438-018-1455-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol. 2005;138(4):2364–73. doi: 10.1104/pp.105.064287 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen A, Dubcovsky J. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012;8(12):e1003134. doi: 10.1371/journal.pgen.1003134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw LM, Lyu B, Turner R, Li C, Chen F, Han X, et al. FLOWERING LOCUS T2 regulates spike development and fertility in temperate cereals. J Exp Bot. 2019;70(1):193–204. doi: 10.1093/jxb/ery350 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Curr Opin Plant Biol. 2009;12(2):178–84. doi: 10.1016/j.pbi.2008.12.010 . [DOI] [PubMed] [Google Scholar]
- 44.Ream TS, Woods DP, Schwartz CJ, Sanabria CP, Mahoy JA, Walters EM, et al. Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol. 2014;164(2):694–709. doi: 10.1104/pp.113.232678 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61(6):1001–13. doi: 10.1111/j.1365-313X.2010.04148.x . [DOI] [PubMed] [Google Scholar]
- 46.Poethig RS. The past, present, and future of vegetative phase change. Plant Physiol. 2010;154(2):541–4. doi: 10.1104/pp.110.161620 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeno K. Stress-induced flowering: the third category of flowering response. J Exp Bot. 2016;67(17):4925–34. doi: 10.1093/jxb/erw272 . [DOI] [PubMed] [Google Scholar]
- 48.Kazan K, Lyons R. The link between flowering time and stress tolerance. J Exp Bot. 2016;67(1):47–60. doi: 10.1093/jxb/erv441 . [DOI] [PubMed] [Google Scholar]
- 49.Cho LH, Yoon J, An G. The control of flowering time by environmental factors. Plant J. 2017;90(4):708–19. doi: 10.1111/tpj.13461 . [DOI] [PubMed] [Google Scholar]
- 50.Shimada S, Ogawa T, Kitagawa S, Suzuki T, Ikari C, Shitsukawa N, et al. A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J. 2009;58(4):668–81: doi: 10.1111/j.1365-313X.2009.03806.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez SE, Kay SA. The plant circadian clock: from a simple timekeeper to a complex developmental manager. Cold Spring Harb Perspect Biol. 2016;8(12). doi: 10.1101/cshperspect.a027748 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379(6):633–46. doi: 10.1515/bchm.1998.379.6.633 . [DOI] [PubMed] [Google Scholar]
- 53.Shigyo M, Hasebe M, Ito M. Molecular evolution of the AP2 subfamily. Gene. 2006;366(2):256–65. doi: 10.1016/j.gene.2005.08.009 . [DOI] [PubMed] [Google Scholar]
- 54.Zhu QH, Helliwell CA. Regulation of flowering time and floral patterning by miR172. J Exp Bot. 2011;62(2):487–95. doi: 10.1093/jxb/erq295 . [DOI] [PubMed] [Google Scholar]
- 55.Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, et al. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell. 2010;22(7):2156–70. doi: 10.1105/tpc.110.075606 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009;7(7):e1000148. doi: 10.1371/journal.pbio.1000148 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang B, Wang L, Zeng L, Zhang C, Ma H. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 2015;29(9):975–87. doi: 10.1101/gad.251520.114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15(11):2730–41. doi: 10.1105/tpc.016238 WOS:000186578900022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, et al. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19(9):2736–48. doi: 10.1105/tpc.107.054528 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lazaro A, Zhou Y, Giesguth M, Nawaz K, Bergonzi S, Pecinka A, et al. PERPETUAL FLOWERING2 coordinates the vernalization response and perennial flowering in Arabis alpina. J Exp Bot. 2019;70(3):949–61. doi: 10.1093/jxb/ery423 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bergonzi S, Albani MC, Ver Loren van Themaat E, Nordström KJ, Wang R, Schneeberger K, et al. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science. 2013;340(6136):1094–7. doi: 10.1126/science.1234116 . [DOI] [PubMed] [Google Scholar]
- 62.Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci U S A. 2005;102(26):9412–7. doi: 10.1073/pnas.0503927102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci U S A. 2007;104(27):11376–81. doi: 10.1073/pnas.0704145104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee YS, Lee DY, Cho LH, An G. Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice (N Y). 2014;7(1):31. doi: 10.1186/s12284-014-0031-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L, Sun S, Jin J, Fu D, Yang X, Weng X, et al. Coordinated regulation of vegetative and reproductive branching in rice. Proc Natl Acad Sci U S A. 2015;112(50):15504–9. doi: 10.1073/pnas.1521949112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Debernardi JM, Lin H, Chuck G, Faris JD, Dubcovsky J. microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development. 2017;144(11):1966–75. doi: 10.1242/dev.146399 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai YS, Gill BS, et al. Molecular characterization of the major wheat domestication gene Q. Genetics. 2006;172(1):547–55. doi: 10.1534/genetics.105.044727 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet. 2007;39(12):1517–21. doi: 10.1038/ng.2007.20 . [DOI] [PubMed] [Google Scholar]
- 69.Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 2009;9:149. doi: 10.1186/1471-2229-9-149 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ó’Maoiléidigh DS, van Driel AD, Singh A, Sang Q, Le Bec N, Vincent C, et al. Systematic analyses of the MIR172 family members of Arabidopsis define their distinct roles in regulation of APETALA2 during floral transition. PLoS Biol. 2021;19(2):e3001043. doi: 10.1371/journal.pbio.3001043 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lian H, Wang L, Ma N, Zhou CM, Han L, Zhang TQ, et al. Redundant and specific roles of individual MIR172 genes in plant development. PLoS Biol. 2021;19(2):e3001044. doi: 10.1371/journal.pbio.3001044 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022–5. doi: 10.1126/science.1088060 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wollmann H, Mica E, Todesco M, Long JA, Weigel D. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development. 2010;137(21):3633–42. doi: 10.1242/dev.036673 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Greenwood JR, Finnegan EJ, Watanabe N, Trevaskis B, Swain SM. New alleles of the wheat domestication gene. Development. 2017;144(11):1959–65. doi: 10.1242/dev.146407 . [DOI] [PubMed] [Google Scholar]
- 75.Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6(7):e1001031. doi: 10.1371/journal.pgen.1001031 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39(4):544–9. doi: 10.1038/ng2001 . [DOI] [PubMed] [Google Scholar]
- 77.Jung JH, Seo PJ, Ahn JH, Park CM. Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J Biol Chem. 2012;287(19):16007–16. doi: 10.1074/jbc.M111.337485 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–9. doi: 10.1016/j.cell.2009.06.031 WOS:000269156100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cardon G, Hohmann S, Klein J, Nettesheim K, Saedler H, Huijser P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene. 1999;237(1):91–104. doi: 10.1016/s0378-1119(99)00308-x WOS:000082500700011. [DOI] [PubMed] [Google Scholar]
- 80.Yang L, Xu M, Koo Y, He J, Poethig RS. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife. 2013;2:e00260. doi: 10.7554/eLife.00260 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu S, Cao L, Zhou CM, Zhang TQ, Lian H, Sun Y, et al. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. Elife. 2013;2:e00269. doi: 10.7554/eLife.00269 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huijser P, Schmid M. The control of developmental phase transitions in plants. Development. 2011;138(19):4117–29. doi: 10.1242/dev.063511 WOS:000294547200005. [DOI] [PubMed] [Google Scholar]
- 83.Wang JW. Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot. 2014;65(17):4723–30. doi: 10.1093/jxb/eru246 . [DOI] [PubMed] [Google Scholar]
- 84.Debernardi JM, Greenwood JR, Jean Finnegan E, Jernstedt J, Dubcovsky J. APETALA 2-like genes AP2L2 and Q specify lemma identity and axillary floral meristem development in wheat. Plant J. 2020;101(1):171–87. doi: 10.1111/tpj.14528 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8(4):517–27. doi: 10.1016/j.devcel.2005.01.018 . [DOI] [PubMed] [Google Scholar]
- 86.Deng W, Casao MC, Wang P, Sato K, Hayes PM, Finnegan EJ, et al. Direct links between the vernalization response and other key traits of cereal crops. Nat Commun. 2015;6:5882. doi: 10.1038/ncomms6882 . [DOI] [PubMed] [Google Scholar]
- 87.Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D, et al. Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc Natl Acad Sci U S A. 2011;108(42):17550–5. doi: 10.1073/pnas.1113971108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu J, Cheng X, Liu P, Sun J. miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 2017;174(3):1931–48. doi: 10.1104/pp.17.00445 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moore I, Gälweiler L, Grosskopf D, Schell J, Palme K. A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci U S A. 1998;95(1):376–81. doi: 10.1073/pnas.95.1.376 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Balanzà V, Martínez-Fernández I, Sato S, Yanofsky MF, Kaufmann K, Angenent GC, et al. Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL-APETALA2 pathway. Nat Commun. 2018;9(1):565. doi: 10.1038/s41467-018-03067-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol. 2006;60(4):469–80. doi: 10.1007/s11103-005-4814-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pearce S, Vanzetti LS, Dubcovsky J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 2013;163(3):1433–45. doi: 10.1104/pp.113.225854 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou Y, Gan X, Viñegra de la Torre N, Neumann U, Albani MC. Beyond flowering time: diverse roles of an APETALA2-like transcription factor in shoot architecture and perennial traits. New Phytol. 2021;229(1):444–59. doi: 10.1111/nph.16839 . [DOI] [PubMed] [Google Scholar]
- 94.Zikhali M, Wingen LU, Leverington-Waite M, Specel S, Griffiths S. The identification of new candidate genes Triticum aestivum FLOWERING LOCUS T3-B1 (TaFT3-B1) and TARGET OF EAT1 (TaTOE1-B1) controlling the short-day photoperiod response in bread wheat. Plant Cell Environ. 2017;40(11):2678–90. doi: 10.1111/pce.13018 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Patil V, McDermott HI, McAllister T, Cummins M, Silva JC, Mollison E, et al. APETALA2 control of barley internode elongation. Development. 2019;146(11):dev170373. doi: 10.1242/dev.170373 WOS:000471800100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–49. doi: 10.1016/j.cell.2009.06.014 WOS:000269156100015. [DOI] [PubMed] [Google Scholar]
- 97.Zhou CM, Zhang TQ, Wang X, Yu S, Lian H, Tang H, et al. Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science. 2013;340(6136):1097–100. doi: 10.1126/science.1234340 . [DOI] [PubMed] [Google Scholar]
- 98.Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev Cell. 2016;37(3):254–66. doi: 10.1016/j.devcel.2016.04.001 . [DOI] [PubMed] [Google Scholar]
- 99.Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell. 2009;17(2):268–78. doi: 10.1016/j.devcel.2009.06.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li C, Lin H, Chen A, Lau M, Jernstedt J, Dubcovsky J. Wheat VRN1, FUL2 and FUL3 play critical and redundant roles in spikelet development and spike determinac. Development. 2019;146(14). doi: 10.1242/dev.175398 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, et al. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115. doi: 10.1186/1471-2229-9-115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39(8):1033–7. doi: 10.1038/ng2079 . [DOI] [PubMed] [Google Scholar]