Figure 2. Core ERQC hubs that control the fate of nascent proteins entering the ER.
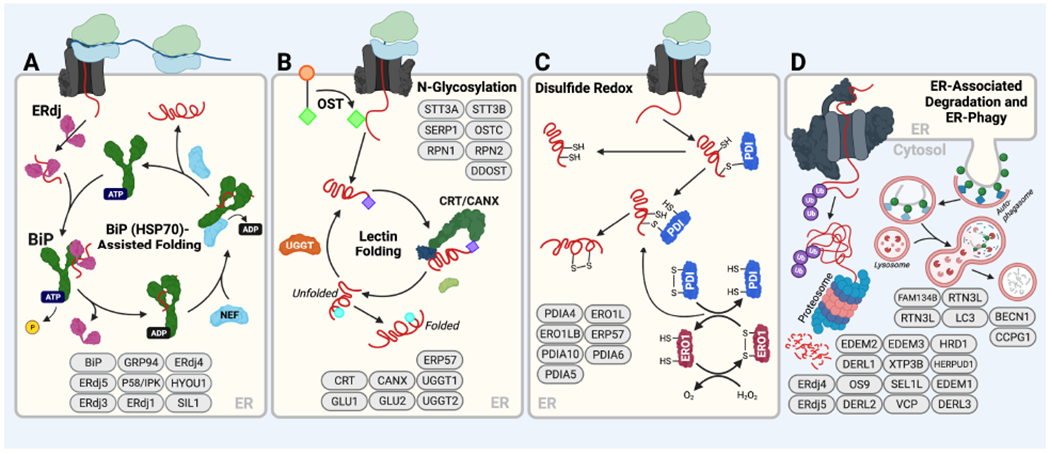
A. The BiP HSP70 chaperoning pathway. Substrates either bind directly to ATP-bound BiP or are delivered by ER-localized J-proteins (ERdjs), which stimulate BiP ATPase activity. This converts BiP to the ADP-bound conformation that has high affinity for substrates. Nucleotide exchange factors (NEFs) such as GRP170/HYOU1 then engage BiP to facilitate ADP-ATP exchange, returning BiP to the low-affinity ATP bound state and releasing the substrate for subsequent rounds of folding. Transfer of a client to ERdj family members like ERdj4 or ERdj5 can remove it from the folding cycle and transfer it for degradation. Components of the BiP chaperoning pathway are shown in ovals at the bottom of the panel. B. The Calnexin/Calreticulin lectin chaperoning pathway. Oligosaccharyl transferase (OST) appends a core glycan comprising Glc3-Man9-GlcNAc2 (green diamonds) to Asn at specific N-glycosyation site sequences (Asn-X-Ser/Thr). Two distal glucose residues are then removed from this core glycan by glucosidases, leaving a protein with a Glc-Man9-GlcNAc2 glycan (purple diamonds). Proteins possessing this singly glucosylated glycan are substrates for the lectin chaperones, calnexin (CANX) and calreticulin (CRT). Upon release from these chaperones, proteins can either fold into their native state or the final glucose of the N-glycan can be removed by glucosidases to leave a glycan comprising Man9-GlcNAc2 (blue circles) that cannot rebind CANX/CRT. This glucose-free glycan can then be either further trimmed by mannosidases that direct the protein to degradation or re-glucosylated by UGGT to allow further rounds of interactions with CANX or CRT. Select components of the Calnexin/Calreticulin lectin chaperoning pathway are shown at the bottom of the panel. C. Protein disulfide isomerase (PDI) activity in the ER. Oxidized PDIs form mixed disulfide bonds with Cys residues in substrate proteins that are then resolved by a second Cys in the substrate, creating a disulfide within the substrate protein. The resulting reduced PDI is then re-oxidized through the activity of ERO1. PDIs like ERdj5, a protein that also includes a J-domain, bind clients while in the reduced state causing the client disulfide to be transferred to ERdj5, thus serving to reduce the client for ERAD. Select components of the PDI folding pathway are shown. D. ER-associated degradation (ERAD) and ER-phagy. In ERAD, terminally misfolded proteins are directed to the ER retrotranslocon, which facilitates removal of non-native proteins from the ER to the cytosol. In the cytosol, these proteins are then ubiquitinated and subsequently degraded by the proteasome. Select components of the ERAD pathway are shown. In ER-phagy, misfolded proteins are directed to the lysosome for degradation by lysosomal hydrolases; select components are shown. Created with BioRender.com.
