Figure 3. Mammalian UPR signaling pathways involved in adapting ERQC following an acute ER insult.
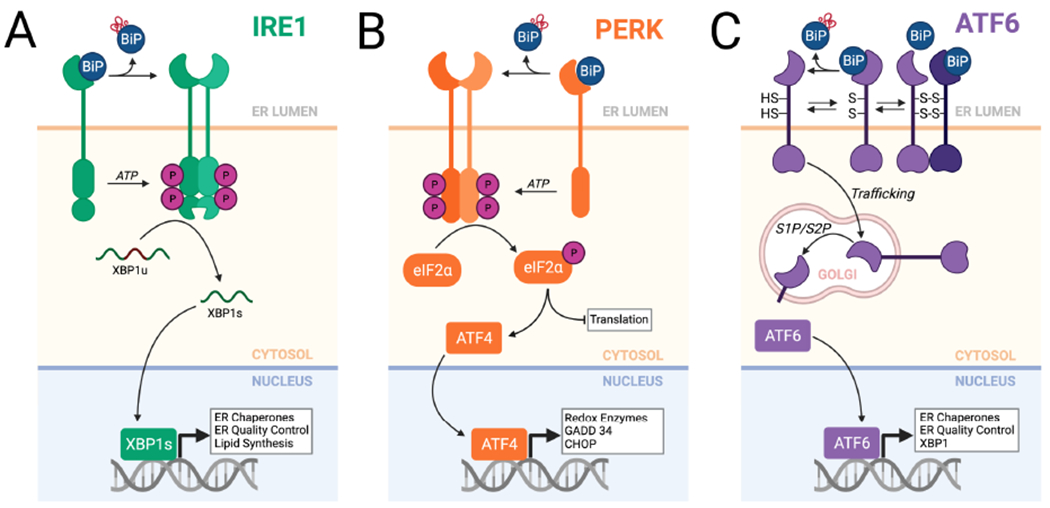
A. The IRE1/XBP1s signaling arm of the UPR. In response to ER stress, IRE1 is activated through a process involving dissociation of BiP from the IRE1 luminal domain. This leads to IRE1 oligomerization and subsequent activation of the IRE1 cytosolic kinase domain to promote autophosphorylation and allosteric activation of the IRE1 RNase. The activated IRE1 RNase adapts ERQC primarily through the non-canonical splicing of XBP1 mRNA, resulting in the production of the active transcription factor XBP1s. XBP1s activates expression of numerous genes involved in ERQC pathways including ER chaperones and degradation factors. B. The PERK arm of the UPR. In response to ER stress, BiP dissociates from the PERK luminal domain, allowing dimerization and autophosphorylation of the cytosolic kinase domain. This leads to selective PERK-dependent phosphorylation of eIF2α, which promotes both translational attenuation and selective activation of stress-responsive transcription factors such as ATF4. C. The ATF6 arm of the UPR. ATF6 is maintained in the ER as monomers and dimers, possessing intra- and inter-molecular disulfide bonds, that are bound to the ER chaperone BiP. ER stress promotes reduction of ATF6 disulfides and BiP dissociation, increasing populations of reduced ATF6 monomers that traffic to the Golgi where they are processed by site 1 (S1P) and site 2 (S2P) proteases. This releases the active ATF6 bZIP transcription factor domain, which localizes to the nucleus and induces expression of stress-responsive genes primarily involved in ERQC. Created with BioRender.com.
