Fig. 5.
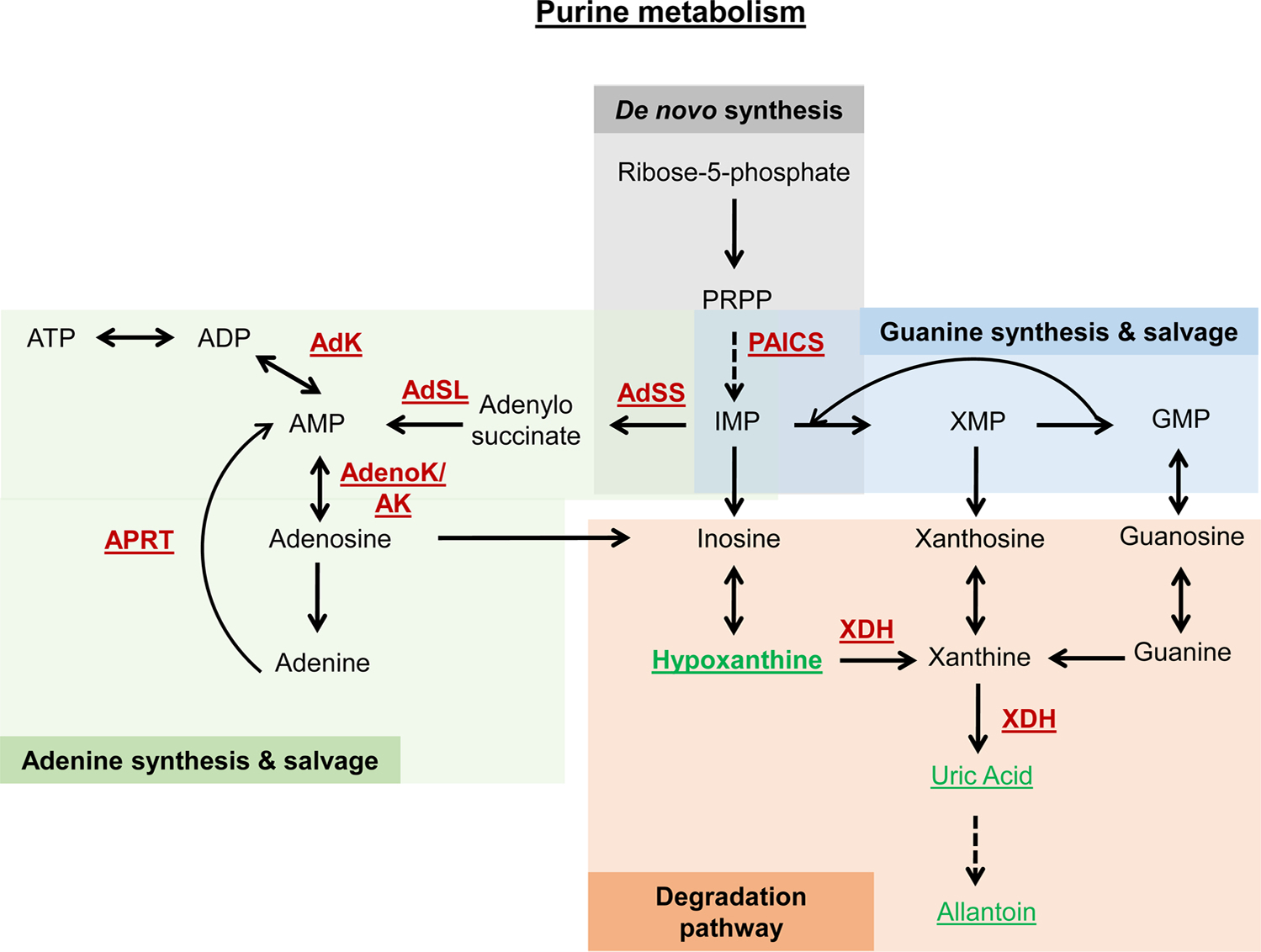
Schematic representation of purine metabolism. Underlined are metabolites and enzymes that were associated with lifespan extension. Red font color represents downregulation or depletion from food, while green font color represents overexpression or supplementation. Dashed line represents that multiple steps are involved. During de novo synthesis, ribose-5-phosphate is converted to inosine monophosphate (IMP), which can then be converted to adenine and guanine nucleotides. In worms, downregulation of PAICS increased lifespan. In flies, heterozygous mutations of adenylosuccinate synthetase (AdSS), adenylosuccinate lyase (AdSL), adenine phosphoribosyltransferase (Aprt), adenosine kinase (AdenoK), and adenylate kinase (AdK) increased lifespan. The degradation pathway converts the purine nucleotides into xanthine, which can then be metabolized to uric acid. Downregulation of xanthine dehydrogenase increased lifespan in flies, while supplementation with hypoxanthine, uric acid and allantoin increased lifespan in worms.
