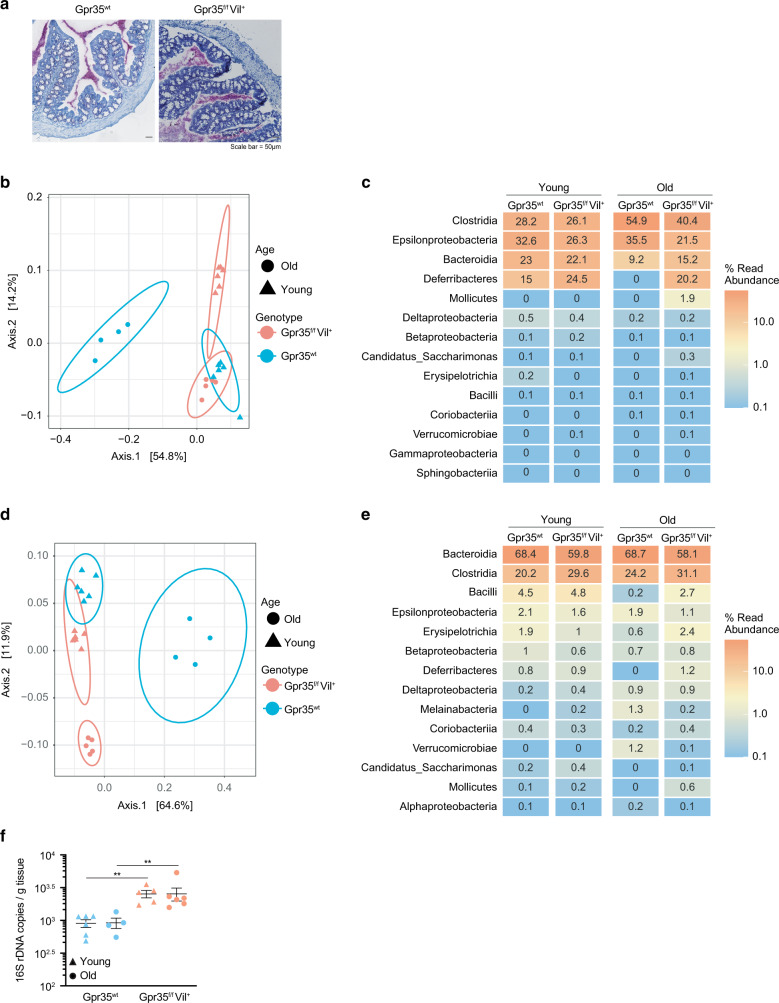
Fig. 2. Epithelial cell-specific Gpr35 deletion correlates with an altered mucosa-associated microbiome.
a Visualization of bacteria in relation to the epithelium via 16S rRNA in situ hybridization (pink) in proximal colon sections obtained from Gpr35f/fVil+ and Gpr35wt co-housed littermates. Scale bars, 50 μm. b Principal component analysis based on Jaccard distance of the rarefied abundance of proximal colon mucosa-associated bacterial communities in Gpr35f/fVil+ young (n = 6), Gpr35f/fVil+ old (n = 5), Gpr35wt young (n = 6) and Gpr35wt old (n = 4) littermates. c Relative abundance of taxonomic groups averaged across mucosa-associated bacteria samples of old and young Gpr35f/fVil+ and Gpr35wt littermates. d Principal component analysis based on Jaccard distance of rarefied abundance of fecal bacterial communities in Gpr35f/fVil+ young (n = 6), Gpr35f/fVil+ old (n = 5), Gpr35wt young (n = 6) and Gpr35wt old (n = 4) littermates. e Relative abundance of taxonomic groups averaged across fecal samples of old and young Gpr35f/fVil+ and Gpr35wt littermates. f 16S rRNA gene qPCR amplification of mucosa-associated bacteria and fecal samples of old and young Gpr35f/fVil+ and Gpr35wt littermates. Fecal and mucosal-associated bacteria were collected from the same animals.