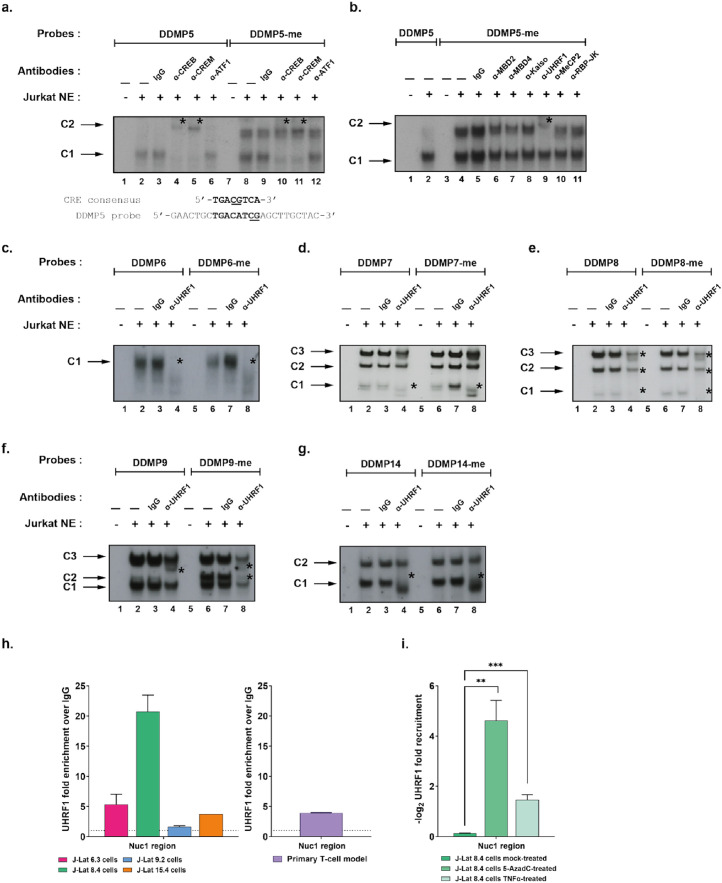
Figure 2.
UHRF1 binds in vitro and in vivo to the latent HIV-1 promoter. (a) The radiolabelled unmethylated or the methylated HIV-1 DDMP5 probe (respectively indicated as “DDMP5” and “DDMP5-me”) were incubated with 10µg of nuclear extracts from Jurkat T cells (“Jurkat NE”) and either with a purified rabbit IgG as a negative control (lane 3 and lane 9) or with an antibody directed against CREB/CREM family members including CREB (lane 4 and lane 10), CREM (lane 5 and lane 11) or ATF1 (lane 6 and lane 12). The figure shows the specific retarded bands of interest indicated by arrows. Supershifted complexes are indicated by asterisks. One representative experiment out of three is presented. (b) The “DDMP5” or “DDMP5-me” probes were incubated with 10 µg of Jurkat cells NE, and either with purified rabbit IgG as a negative control (lane 5) or with an antibody directed against methylcytosines-recognizing proteins, including MBD2 (lane 6), MBD4 (lane 7), Kaiso (lane 8), UHRF1 (lane 9), MeCP2 (lane 10) and RBP-JK (lane 11). The figure shows the specific retarded bands of interest indicated by arrows. Supershifted complexes are indicated by asterisks. One representative experiment out of three is presented. (c, d, e, f, g) The “DDMP6” or “DDMP6-me” probes (c), “DDMP7” or “DDMP7-me” probes (d), “DDMP8” or “DDMP8-me” probes (e), “DDMP9” or “DDMP9-me” probes (f) and “DDMP14” or “DDMP14-me” probes (g) were incubated with 10µg of Jurkat cells NE, and either with purified rabbit IgG as a negative control (lanes 3 and 7) or antibodies directed against UHRF1 (lanes 4 and 8). The figure shows the specific retarded bands of interest indicated by arrows. Supershifted complexes are indicated by asterisks. One representative experiment out of three is presented. (h) Chromatin preparations of J-Lat cells and primary CD4+ T cell models for HIV-1 latency were immunoprecipitated with an anti-UHRF1 antibody or with purified rabbit IgG, serving as a negative control. qPCRs were performed with primers hybridizing specifically to the 5’LTR, in the Nuc-1 region. Folds relative to IgG are presented, where fold enrichments for each immunoprecipitated DNA were calculated by the relative standard curve on input DNA. Values represent the means of duplicate samples ± SD. One representative experiment out of three is presented, except for the primary CD4+ T cell model which is representative of two independent infections of healthy donors. (i) Chromatin preparations of J-Lat 8.4 cells, either mock-treated or treated with 400 nM of 5-AzadC or with 10 ng/mL of TNF-α, were immunoprecipitated with an anti-UHRF1 antibody or with purified rabbit IgG, serving as a negative control. Folds relative to IgG were first calculated as above, then normalized to the mock condition and -log2(fold to mock) were plotted. Statistical significance was calculated with an [unpaired T-test].