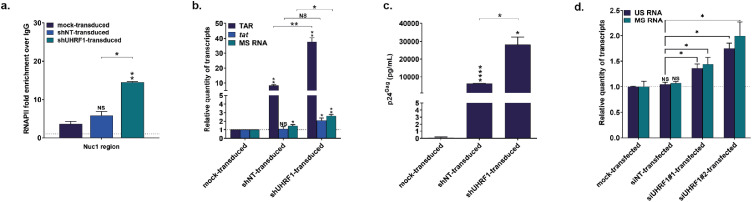
Figure 3.
UHRF1 silences HIV-1 transcription during latency. (a) Chromatin was prepared from J-Lat 8.4 cells, either mock-transduced, control-transduced (with non-targeting shRNA, indicated as “shNT-transduced”) or shUHRF1-transduced (indicated as “shUHRF1-transduced”). Immunoprecipitations were performed using anti-RNAPII or purified rabbit IgG, as a negative control. qPCRs were performed with primers hybridizing specifically to the 5’LTR, in the Nuc-1 region. Folds relative to IgG are presented, where fold enrichments for each immunoprecipitated DNA were calculated by the relative standard curve on input DNA. Values represent the means of duplicate samples ± SD. (b) Total RNA preparations from mock-transduced, shNT-transduced or shUHRF1-transduced J-Lat 8.4 cells were reverse transcribed. Initiated (TAR region), elongated (tat region) transcripts, or HIV-1 multiply spliced RNA (MS RNA) were quantified by RT-qPCR using GAPDH as a first normalizer and the mock-transduced condition as a second normalizer. Means from duplicate ± SD are indicated. Statistical significance was calculated with an unpaired T test. (c) Cultures supernatants from mock-transduced, shNT-transduced or shUHRF1-transduced J-Lat 8.4 cells were probed for viral production as measured by ELISA on p24Gag capsid protein. (d) Total RNA preparations from mock-transfected, siNT-transfected, siUHRF1#1-transfected or siUHRF1#1 primary CD4+ T cell models were reverse transcribed. Unspliced HIV-1 RNA (US RNA) or multiply spliced HIV-1 RNA (MS RNA) were quantified by RT-qPCR using TBP as a first normalizer and the mock-transfected condition as a second normalizer. Means from duplicate ± SD are indicated. One representative model out of two is shown. Statistical significance was calculated with a Mann-Whitney test.