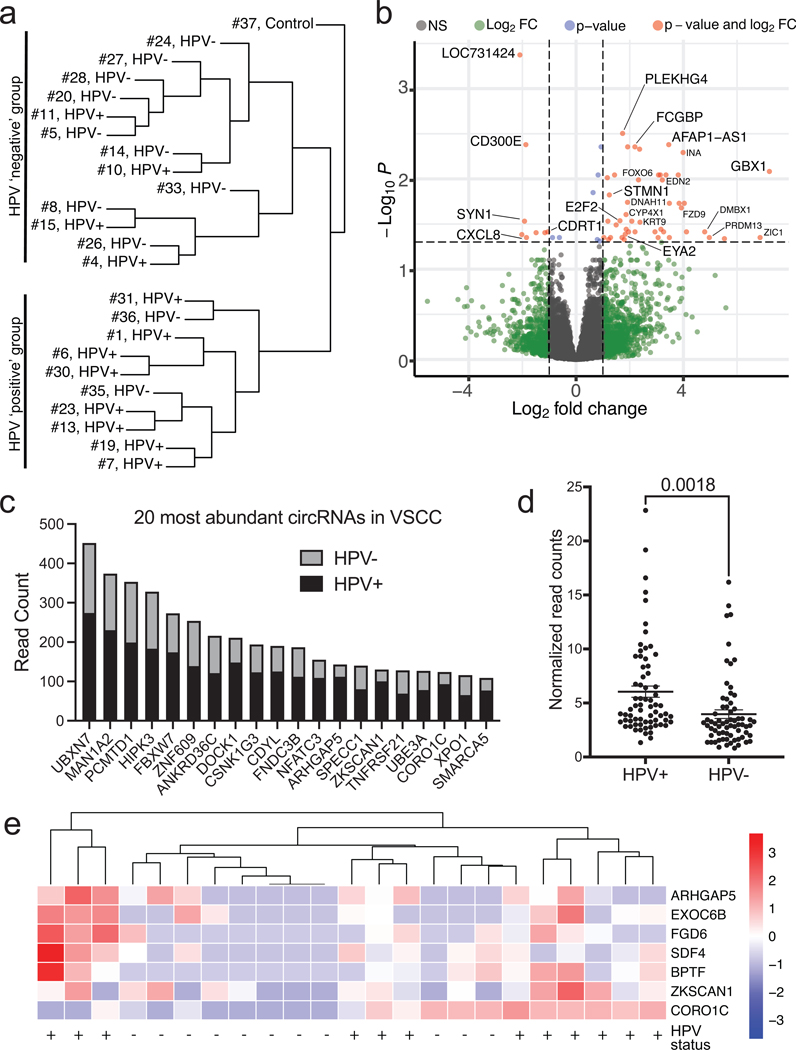
Figure 3. RNA-Sequencing Analysis of Vulvar Squamous Cell Carcinoma.
a. Unsupervised hierarchical clustering broken down into HPV-positive and HPV-negative cases. Samples cluster based upon their HPV status.
b. Differential gene expression of HPV-positive vs. HPV-negative illustrated using a volcano plot.
c. Bar graph detailing the 20 most abundant circular RNAs found in VSCC determined by read count. Each bar is broken down into the sum of read counts based from HPV-positive or HPV-negative.
d. Plots of circRNA read counts normalized by sample number. Each dot represents a distinct circRNA with >50 total read counts. HPV-positive samples have significantly higher circRNA read counts.
e. Scaled heatmap demonstrating differential circRNAs in HPV-positive or HPV-negative samples.