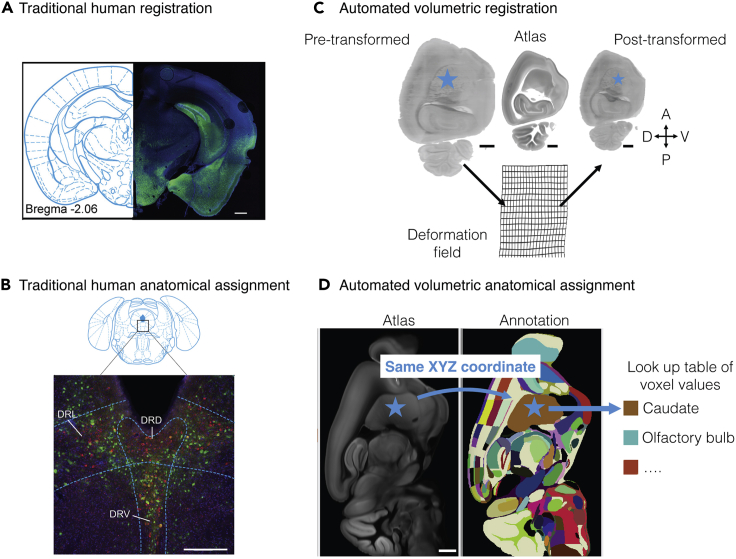
Figure 9.
Methodological comparison of traditional anatomical assignment with automated anatomical assignment
(A and B) Traditional manual-based tissue-section registration and subregional anatomical assignment example. Modified from Pomeranz et al. (2017). (A) Gross cytoarchitecture and tissue-section shape is used for traditional visual registration. Human-user visually defined coronal section atlas correspondence of fluorescent tissue after PRV injection. (B) Cytoarchitecture can be used for traditional anatomical assignment. After section registration, subregional anatomical assignment is hand-drawn.
(C and D) Automated registration and subregional assignment. The blue star illustrates how a point (cell center) can be transformed into different space for eventual anatomical assignment. (C) Volumetric registration transforms light-sheet volumes into atlas space. Automated mapping between the pre-transformed volume and the atlas is established. Deformation fields illustrate two-dimensional plane deformation. (D) Anatomical assignment is automated after volumetric registration to atlas space. The pixel value in annotation space of voxel-coordinates is an identity lookup table and can be used to determine region identity. Scale bars are 500 μm (A), 250 μm (B), 1 mm (C and D). Abbreviations: DRD, dorsomedial raphe nucleus; DRL, dorsolateral raphe nucleus, DRV, ventromedial raphe nucleus.