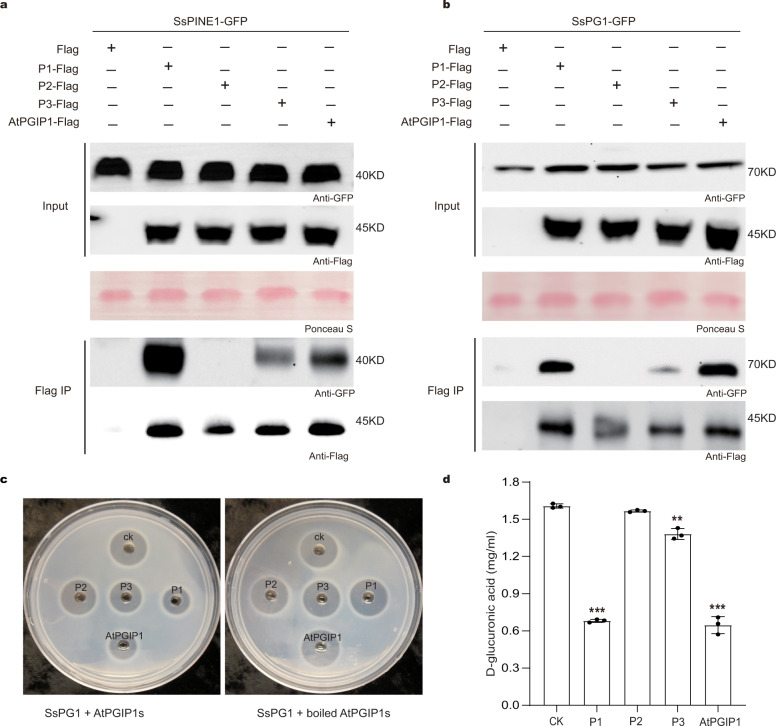
Fig. 5. The P2 region of AtPGIP1 determines its binding specificity with SsPINE1 and SsPG1.
a Co-IP assays showing the AtPGIP1 variants P2 and P3 have lost and reduced, respectively, affinity with SsPINE1. SsPINE1-GFP was co-expressed with 3xFlag-tagged AtPGIP1, AtPGIP1-P1, AtPGIP1-P2, or AtPGIP1-P3 in N. benthamiana leaves. The samples were then used for Flag IPs and anti-GFP Western blots. b Similar Co-IP assays were carried out to test the bindings of SsPG1-GFP to 3xFlag-tagged AtPGIP1, AtPGIP1-P1, AtPGIP1-P2, or AtPGIP1-P3. Co-IP assays showing the AtPGIP1 variants P2 and P3 have lost and reduced affinity with SsPG1, respectively. SsPG1-GFP was co-expressed with 3xFlag-tagged AtPGIP1, AtPGIP1-P1, AtPGIP1-P2, or AtPGIP1-P3 in N. benthamiana leaves. The samples were then used for Flag IPs and anti-GFP Western blots. c Agar diffusion assay showing the SsPG1-inhibitory activity of AtPGIP1 and its mutation variants P1, P2 and P3. The wells were applied with 0.5 μg of SsPG1 alone (CK) and with 10 μg of either variant P1, P2, P3, or wildtype AtPGIP1 proteins. d PG activity of SsPG1 alone (CK) and in the presence of AtPGIP1 or its mutation variants were determined by measuring the released D-galacturonic acid. Data represent means ± s.d. of n = 3 independent replicates. ** and *** indicate significant difference from the check (CK, SsPG1 alone) at p < 0.01 and 0.001, respectively, in two-tailed t-test. Source data are provided as a Source data file.